Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC010426A_C01 KMC010426A_c01
(573 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
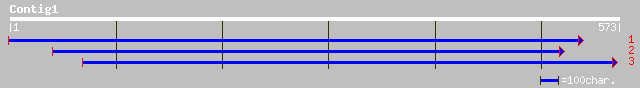
ref|NP_602507.1| Hypothetical membrane-spanning protein [Fusobac... 35 0.67
gb|EAA20829.1| ATP-dependent RNA helicase [Plasmodium yoelii yoe... 33 1.9
gb|EAA20911.1| clp [Plasmodium yoelii yoelii] 33 3.3
gb|EAA16805.1| hypothetical protein [Plasmodium yoelii yoelii] 32 7.4
ref|NP_473027.1| hypothetical protein [Plasmodium falciparum 3D7... 32 7.4
>ref|NP_602507.1| Hypothetical membrane-spanning protein [Fusobacterium nucleatum
subsp. nucleatum ATCC 25586] gi|19712922|gb|AAL93806.1|
Hypothetical membrane-spanning protein [Fusobacterium
nucleatum subsp. nucleatum ATCC 25586]
Length = 382
Score = 35.0 bits (79), Expect = 0.67
Identities = 17/63 (26%), Positives = 41/63 (64%), Gaps = 3/63 (4%)
Frame = -3
Query: 487 VIIDGNYH---FIAFYSLHVVFFFRIRCKLVVLMQIVSYLFKSINLMLLVTFLLQVKMNF 317
V++ G +H F+A + L+ +F ++I+ K ++++ ++ ++FK +L F+L+ K+++
Sbjct: 151 VLLGGLFHKSLFLAIF-LYPIFKYKIKDKYLIMIFLILFVFKRELFEVLEFFILKYKISY 209
Query: 316 ESY 308
SY
Sbjct: 210 SSY 212
>gb|EAA20829.1| ATP-dependent RNA helicase [Plasmodium yoelii yoelii]
Length = 867
Score = 33.5 bits (75), Expect = 1.9
Identities = 15/39 (38%), Positives = 25/39 (63%)
Frame = +2
Query: 29 NRYMNNIKIGHPSLLNTKPNNVINSLKHHK*QNLQHILT 145
N +NNI HP++L + P ++ N + HK +N Q+IL+
Sbjct: 217 NHNVNNIFYDHPTILVSTPKHLCNYILEHKNKNNQNILS 255
>gb|EAA20911.1| clp [Plasmodium yoelii yoelii]
Length = 518
Score = 32.7 bits (73), Expect = 3.3
Identities = 30/141 (21%), Positives = 70/141 (49%), Gaps = 6/141 (4%)
Frame = -3
Query: 490 NVIIDGNYHFIAFYSLHVVF-FFRIR--CKLV---VLMQIVSYLFKSINLMLLVTFLLQV 329
N +++ N+ ++V+F FF+IR CK + ++++I+ YLF IN +L + +
Sbjct: 342 NNVMEDNFLINISKIVNVIFDFFQIRDLCKSIHKDIILRIIDYLFYHINQYILDYIIEKT 401
Query: 328 KMNFESYSQQNLTPMKRMSHQNLVYSDIAENYPAQASSTKELKRVKIEK*NKNLITLRLL 149
K+N + + + + + + + D +N Q ++ E + ++ + + L L
Sbjct: 402 KLNDDERNSIHEKFIFIQNSLSFILEDYKDNEFEQKNNIVEEENTYLKIAGDDPVCLISL 461
Query: 148 LR*YMLQVLLFMVF*RVNYII 86
+ +VL ++F ++ YI+
Sbjct: 462 KK---NRVLFLLLFCKIKYIL 479
>gb|EAA16805.1| hypothetical protein [Plasmodium yoelii yoelii]
Length = 955
Score = 31.6 bits (70), Expect = 7.4
Identities = 30/123 (24%), Positives = 58/123 (46%), Gaps = 4/123 (3%)
Frame = -3
Query: 514 SVNPNIILNVIIDGNYHFIAFYSLHVVFFFRIRCKLVVLMQIVSYLFKSINLMLLVTFLL 335
+VN I +N ++ N F+ ++ + F + KL + + + + +I + L + +
Sbjct: 665 NVNNIIQINKLLYKNNIFLKIFNFYE--FLTLPSKLTRIQILYNIIKSNIQMNLNYSLID 722
Query: 334 QVKMNFESYSQQNLTP---MKRMSHQNLVYSDIAENYPAQASSTKELKRVKIEK*N-KNL 167
Q+ ++ NL KR++ +L S AEN +A S + K++KI N KN
Sbjct: 723 QIYNELYLFTTNNLRNKHFQKRIACNHLSVSSNAENEANKAESETDYKKIKIININGKNK 782
Query: 166 ITL 158
I +
Sbjct: 783 IVV 785
>ref|NP_473027.1| hypothetical protein [Plasmodium falciparum 3D7]
gi|7494297|pir||F71613 hypothetical protein PFB0495w -
malaria parasite (Plasmodium falciparum)
gi|3845199|gb|AAC71888.1| hypothetical protein
[Plasmodium falciparum 3D7]
Length = 1121
Score = 31.6 bits (70), Expect = 7.4
Identities = 36/135 (26%), Positives = 63/135 (46%), Gaps = 12/135 (8%)
Frame = -3
Query: 520 LTSVNPNIILNVIIDGNYHFIAFYS------LHVVFF-FRIRCKLVVLMQIVSYLFKSIN 362
L S+N ILN IID N FI FY L++ F F+ C ++ ++ + Y N
Sbjct: 386 LNSLNNKFILNKIIDKN--FILFYECLLKILLNIKFVNFQSLCISLISLKNIYYNILRNN 443
Query: 361 LMLLVTFLLQVKMNFESYSQQNLTPMKRMSHQN-----LVYSDIAENYPAQASSTKELKR 197
+ ++ L M F Y N+ KR+ +N +++++ NY S+ + +K
Sbjct: 444 VYIVNNVLFNDIMKFSLY-LCNIFLGKRIKTENENAVLIIHNNDQTNY----SNKENIKD 498
Query: 196 VKIEK*NKNLITLRL 152
+ I+K K I ++
Sbjct: 499 IIIQKRIKEYIFYKM 513
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 384,375,111
Number of Sequences: 1393205
Number of extensions: 6720583
Number of successful extensions: 12955
Number of sequences better than 10.0: 12
Number of HSP's better than 10.0 without gapping: 12568
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 12945
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 21243732558
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)