Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC010311A_C01 KMC010311A_c01
(541 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
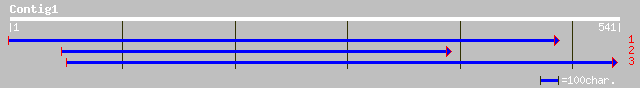
ref|NP_181580.1| hypothetical protein; protein id: At2g40480.1 [... 53 2e-06
dbj|BAC04594.1| unnamed protein product [Homo sapiens] 34 1.3
dbj|BAC05810.1| seven transmembrane helix receptor [Homo sapiens] 34 1.3
ref|NP_459965.1| putative KicA protein [Salmonella typhimurium L... 32 4.9
ref|NP_455476.1| putative membrane protein [Salmonella enterica ... 32 4.9
>ref|NP_181580.1| hypothetical protein; protein id: At2g40480.1 [Arabidopsis
thaliana] gi|25408707|pir||A84830 hypothetical protein
At2g40480 [imported] - Arabidopsis thaliana
gi|2651308|gb|AAB87588.1| hypothetical protein
[Arabidopsis thaliana]
Length = 541
Score = 53.1 bits (126), Expect = 2e-06
Identities = 35/86 (40%), Positives = 54/86 (62%), Gaps = 7/86 (8%)
Frame = -1
Query: 541 MRDVLSRKQVP-EEYTATKEMEERGE---SNVALSPMLRALREDPTPTAQHDHDR-SDQQ 377
MRDVL RKQVP E+ A + G+ NV LS ML+ L++D +A+ + + +++
Sbjct: 454 MRDVLRRKQVPKEDVVAPQRQSLEGQIPRRNVNLSQMLKELKQDVKFSARGEKEEVHEEK 513
Query: 376 QFT--RKKFGFIPISLPLGKRRNKRA 305
Q+ R+KFGFI I+LPL K+ K++
Sbjct: 514 QYVTQRRKFGFIHITLPLQKQSKKKS 539
>dbj|BAC04594.1| unnamed protein product [Homo sapiens]
Length = 134
Score = 33.9 bits (76), Expect = 1.3
Identities = 20/75 (26%), Positives = 32/75 (42%)
Frame = -2
Query: 231 CLAILY*FLCYVSLFASCVEVCVGV*FCEICFCFVIGLHYATSLALMRLMNLLLCMKTGV 52
CL + ++C CV VCV V C IC C + + + + + +C+ V
Sbjct: 14 CLCVCI-YMCVCVPVCMCVYVCVYVRICVICVCVCVFVCAHMCSCVFVYVCICVCLYVCV 72
Query: 51 FNVVLYYVKFYMDIC 7
F V YV + +C
Sbjct: 73 FVCVCMYVYLCVFVC 87
>dbj|BAC05810.1| seven transmembrane helix receptor [Homo sapiens]
Length = 346
Score = 33.9 bits (76), Expect = 1.3
Identities = 19/76 (25%), Positives = 36/76 (47%), Gaps = 1/76 (1%)
Frame = -2
Query: 231 CLAILY*FLCYVSLFASCVEVCVGV*FC-EICFCFVIGLHYATSLALMRLMNLLLCMKTG 55
CL + Y+ ++ C+ VCV V C +C C + + S+ L + + +C+
Sbjct: 106 CLCVYLHISVYLCVYV-CICVCVSVSLCVSVCACLSLCVCIRVSVCLSLRVCVCVCLCVS 164
Query: 54 VFNVVLYYVKFYMDIC 7
V+ V Y+ Y+ +C
Sbjct: 165 VYVCVSSYLCVYLCVC 180
Score = 33.1 bits (74), Expect = 2.2
Identities = 19/82 (23%), Positives = 36/82 (43%), Gaps = 11/82 (13%)
Frame = -2
Query: 219 LY*FLCYVSLFASCVEVCVGV*FCE-----------ICFCFVIGLHYATSLALMRLMNLL 73
LY ++C + CV +CV V C IC C + + S+++ + L
Sbjct: 17 LYLYVCVCICVSVCVCICVSVCVCICVCVSVFVCVCICVCVCVCVSMCVSISVCVYLCLC 76
Query: 72 LCMKTGVFNVVLYYVKFYMDIC 7
+C+ V+ V Y+ ++ +C
Sbjct: 77 VCVSVCVYVSVCMYLCVFLCVC 98
>ref|NP_459965.1| putative KicA protein [Salmonella typhimurium LT2]
gi|16419502|gb|AAL19924.1| putative KicA protein
[Salmonella typhimurium LT2]
Length = 258
Score = 32.0 bits (71), Expect = 4.9
Identities = 15/32 (46%), Positives = 19/32 (58%)
Frame = +1
Query: 400 HVGLWELDLPSRLEALDSMLRCFHRALPSPWW 495
H GL L P+ A+DS L + RA+PSP W
Sbjct: 199 HAGLNPLPAPANQLAIDSPLNPWERAIPSPVW 230
>ref|NP_455476.1| putative membrane protein [Salmonella enterica subsp. enterica
serovar Typhi] gi|29142368|ref|NP_805710.1| putative
membrane protein [Salmonella enterica subsp. enterica
serovar Typhi Ty2] gi|25347649|pir||AD0615 probable
membrane protein STY0992 [imported] - Salmonella
enterica subsp. enterica serovar Typhi (strain CT18)
gi|16502152|emb|CAD05390.1| putative membrane protein
[Salmonella enterica subsp. enterica serovar Typhi]
gi|29137998|gb|AAO69559.1| putative membrane protein
[Salmonella enterica subsp. enterica serovar Typhi Ty2]
Length = 258
Score = 32.0 bits (71), Expect = 4.9
Identities = 15/32 (46%), Positives = 19/32 (58%)
Frame = +1
Query: 400 HVGLWELDLPSRLEALDSMLRCFHRALPSPWW 495
H GL L P+ A+DS L + RA+PSP W
Sbjct: 199 HAGLNPLPAPANQLAIDSPLNPWERAIPSPVW 230
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 457,832,341
Number of Sequences: 1393205
Number of extensions: 9711952
Number of successful extensions: 23345
Number of sequences better than 10.0: 11
Number of HSP's better than 10.0 without gapping: 22681
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 23335
length of database: 448,689,247
effective HSP length: 115
effective length of database: 288,470,672
effective search space used: 18462123008
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)