Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC010177A_C01 KMC010177A_c01
(692 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
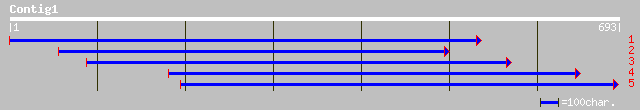
ref|NP_194153.1| putative protein; protein id: At4g24220.1, supp... 174 1e-42
gb|AAO63776.1| unknown [Populus tremuloides] 170 2e-41
emb|CAC80137.1| progesterone 5-beta-reductase [Digitalis purpurea] 165 6e-40
dbj|BAC20032.1| putative induced upon wounding stress [Oryza sat... 137 2e-31
emb|CAA68126.1| induced upon wounding stress [Arabidopsis thaliana] 137 2e-31
>ref|NP_194153.1| putative protein; protein id: At4g24220.1, supported by cDNA:
34176., supported by cDNA: gi_17064749 [Arabidopsis
thaliana] gi|7487331|pir||T09885 hypothetical protein
T22A6.50 - Arabidopsis thaliana
gi|5051764|emb|CAB45057.1| putative protein [Arabidopsis
thaliana] gi|7269272|emb|CAB79332.1| putative protein
[Arabidopsis thaliana] gi|17064750|gb|AAL32529.1|
putative protein [Arabidopsis thaliana]
gi|21592923|gb|AAM64873.1| induced upon wounding stress
[Arabidopsis thaliana]
Length = 388
Score = 174 bits (440), Expect = 1e-42
Identities = 81/119 (68%), Positives = 95/119 (79%)
Frame = -3
Query: 690 AFNCSNGDVFKWKHLWKVLAEQFGIEEYGLFEEDSSGWRLSEMMKDKGAVWEEIVKENQL 511
AFNC+N D+FKWKHLWK+LAEQFGIEEYG E + G L EMMK K VWEE+VKENQL
Sbjct: 272 AFNCNNADIFKWKHLWKILAEQFGIEEYGFEEGKNLG--LVEMMKGKERVWEEMVKENQL 329
Query: 510 QPTKLHEVAEWWFVELMFSGGGLLDSMNKAKEHGFLGFRNTKNSFISWIDKTKAYKIVP 334
Q KL EV WWF +++ G++DSMNK+KE+GFLGFRN+ NSFISWIDK KA+KIVP
Sbjct: 330 QEKKLEEVGVWWFADVILGVEGMIDSMNKSKEYGFLGFRNSNNSFISWIDKYKAFKIVP 388
>gb|AAO63776.1| unknown [Populus tremuloides]
Length = 391
Score = 170 bits (430), Expect = 2e-41
Identities = 78/119 (65%), Positives = 95/119 (79%)
Frame = -3
Query: 690 AFNCSNGDVFKWKHLWKVLAEQFGIEEYGLFEEDSSGWRLSEMMKDKGAVWEEIVKENQL 511
AFN +NGD+FKWKH+W VLA++FGIE+YG E +SS +E MKDKG VWEEIVK+NQL
Sbjct: 274 AFNINNGDLFKWKHMWTVLAQEFGIEKYGFVEGESSV-TFAEKMKDKGPVWEEIVKKNQL 332
Query: 510 QPTKLHEVAEWWFVELMFSGGGLLDSMNKAKEHGFLGFRNTKNSFISWIDKTKAYKIVP 334
KL +V WWF +L+FSG G + SMNKAKEHGFLGFRN+K SF+SWI K +AYK+VP
Sbjct: 333 LSNKLEQVGGWWFGDLIFSGSGYVASMNKAKEHGFLGFRNSKKSFVSWIHKMRAYKVVP 391
>emb|CAC80137.1| progesterone 5-beta-reductase [Digitalis purpurea]
Length = 389
Score = 165 bits (417), Expect = 6e-40
Identities = 79/121 (65%), Positives = 93/121 (76%), Gaps = 2/121 (1%)
Frame = -3
Query: 690 AFNCSNGDVFKWKHLWKVLAEQFGIE--EYGLFEEDSSGWRLSEMMKDKGAVWEEIVKEN 517
AFN SNGDVFKWKH WKVLAEQFG+E EYG + +L ++MK K AVWEEIV+EN
Sbjct: 273 AFNVSNGDVFKWKHFWKVLAEQFGVECGEYG----EGVDLKLQDLMKGKEAVWEEIVREN 328
Query: 516 QLQPTKLHEVAEWWFVELMFSGGGLLDSMNKAKEHGFLGFRNTKNSFISWIDKTKAYKIV 337
L PTKL ++ WWF +++ LDSMNK+KEHGFLGFRN+KN+FISWIDK KAYKIV
Sbjct: 329 GLTPTKLKDIGIWWFGDVILGNECFLDSMNKSKEHGFLGFRNSKNAFISWIDKAKAYKIV 388
Query: 336 P 334
P
Sbjct: 389 P 389
>dbj|BAC20032.1| putative induced upon wounding stress [Oryza sativa (japonica
cultivar-group)]
Length = 410
Score = 137 bits (344), Expect = 2e-31
Identities = 68/120 (56%), Positives = 84/120 (69%), Gaps = 1/120 (0%)
Frame = -3
Query: 690 AFNCSNGDVFKWKHLWKVLAEQFGIEEYGLFEEDSSGWRLSEMMKDKGAVWEEIVKENQL 511
AFNCSNGD++KWK LW VLA +FG+E G +E + L+ M K AVW EIV E +L
Sbjct: 292 AFNCSNGDIYKWKQLWPVLAGKFGVEWAG-YEGEERRVGLTAAMAGKEAVWAEIVAEEKL 350
Query: 510 QPTKLHEVAEWWFVELMFSGGG-LLDSMNKAKEHGFLGFRNTKNSFISWIDKTKAYKIVP 334
T+L EVA WWFV+ +F +D+MNK+KEHGFLGFRNT SF +WIDK K Y+IVP
Sbjct: 351 VATELGEVANWWFVDALFMDKWEFVDTMNKSKEHGFLGFRNTLRSFEAWIDKMKLYRIVP 410
>emb|CAA68126.1| induced upon wounding stress [Arabidopsis thaliana]
Length = 386
Score = 137 bits (344), Expect = 2e-31
Identities = 67/103 (65%), Positives = 77/103 (74%)
Frame = -3
Query: 642 KVLAEQFGIEEYGLFEEDSSGWRLSEMMKDKGAVWEEIVKENQLQPTKLHEVAEWWFVEL 463
K+LAEQFGIEEYG W L EMMK K VWEE+VKENQLQ KL EV WWF ++
Sbjct: 289 KILAEQFGIEEYG-----GRIWGLVEMMKGKERVWEEMVKENQLQEKKLEEVGVWWFADV 343
Query: 462 MFSGGGLLDSMNKAKEHGFLGFRNTKNSFISWIDKTKAYKIVP 334
+ G++DSMNK KE+GFLGFRN+ NSFISWIDK KA+KIVP
Sbjct: 344 ILGVEGMIDSMNKRKEYGFLGFRNSNNSFISWIDKYKAFKIVP 386
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 643,535,956
Number of Sequences: 1393205
Number of extensions: 15028694
Number of successful extensions: 56694
Number of sequences better than 10.0: 108
Number of HSP's better than 10.0 without gapping: 50901
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 56091
length of database: 448,689,247
effective HSP length: 119
effective length of database: 282,897,852
effective search space used: 31401661572
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)