Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
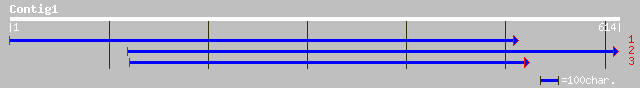
Query= KMC010169A_C01 KMC010169A_c01
(495 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAN17417.1| mRNA capping enzyme - like protein [Arabidopsis t... 40 0.011
ref|NP_195749.1| mRNA capping enzyme - like protein; protein id:... 40 0.011
ref|NP_187522.1| putative mRNA capping enzyme, RNA guanylyltrans... 40 0.019
ref|NP_078224.1| conserved hypothetical ATP/GTP-binding protein ... 38 0.070
gb|AAL92380.1| hypothetical protein [Dictyostelium discoideum] g... 31 6.6
>gb|AAN17417.1| mRNA capping enzyme - like protein [Arabidopsis thaliana]
gi|24899669|gb|AAN65049.1| mRNA capping enzyme - like
protein [Arabidopsis thaliana]
Length = 657
Score = 40.4 bits (93), Expect = 0.011
Identities = 19/28 (67%), Positives = 21/28 (74%)
Frame = -2
Query: 380 FWLLSTVTKLLKEFIKRLSHETNGLYFR 297
FWLLSTV KLLK I LSHE +GL F+
Sbjct: 486 FWLLSTVEKLLKNTIPSLSHEADGLIFQ 513
>ref|NP_195749.1| mRNA capping enzyme - like protein; protein id: At5g01290.1
[Arabidopsis thaliana] gi|11358515|pir||T45969 mRNA
capping enzyme-like protein - Arabidopsis thaliana
gi|6759452|emb|CAB69857.1| mRNA capping enzyme-like
protein [Arabidopsis thaliana]
Length = 607
Score = 40.4 bits (93), Expect = 0.011
Identities = 19/28 (67%), Positives = 21/28 (74%)
Frame = -2
Query: 380 FWLLSTVTKLLKEFIKRLSHETNGLYFR 297
FWLLSTV KLLK I LSHE +GL F+
Sbjct: 436 FWLLSTVEKLLKNTIPSLSHEADGLIFQ 463
>ref|NP_187522.1| putative mRNA capping enzyme, RNA guanylyltransferase; protein id:
At3g09100.1 [Arabidopsis thaliana]
gi|5923675|gb|AAD56326.1|AC009326_13 putative mRNA
capping enzyme, RNA guanylyltransferase [Arabidopsis
thaliana]
Length = 653
Score = 39.7 bits (91), Expect = 0.019
Identities = 18/28 (64%), Positives = 21/28 (74%)
Frame = -2
Query: 380 FWLLSTVTKLLKEFIKRLSHETNGLYFR 297
FWLLS V K+LK FI LSHE +GL F+
Sbjct: 491 FWLLSAVEKVLKGFIPSLSHEADGLIFQ 518
>ref|NP_078224.1| conserved hypothetical ATP/GTP-binding protein [Ureaplasma
urealyticum] gi|11356758|pir||G82897 conserved
hypothetical ATP/GTP-binding protein UU389 [imported] -
Ureaplasma urealyticum
gi|6899374|gb|AAF30799.1|AE002136_6 conserved
hypothetical ATP/GTP-binding protein [Ureaplasma
urealyticum]
Length = 1284
Score = 37.7 bits (86), Expect = 0.070
Identities = 35/130 (26%), Positives = 56/130 (42%), Gaps = 17/130 (13%)
Frame = +1
Query: 100 QAFYDLK*KYNNQKKYNNYIRSHHICSQCSNHSVDISL---ALRW*PNFLP*V-LLSPFI 267
+A YD KYN + NN+++ Q + +D +L +L FLP + LL F+
Sbjct: 528 KASYDFFKKYNYEFSKNNFLKFKEESEQLNKLQIDFNLITKSLYQSQTFLPRIKLLESFL 587
Query: 268 EKK*LTIT------INLKYRPFVSCDSLFMN-------SFKSFVTVESNQKNLSSPSKHL 408
K +T+T +NL +V + + N FK F +E N K+ S HL
Sbjct: 588 VKNNMTLTSLIYDDVNLLSNTYVEANEFYTNFNHLLKDDFKKF--IEDNIKSHSLFFSHL 645
Query: 409 GIPHSTIWAF 438
+ W +
Sbjct: 646 NTLKNNEWTY 655
>gb|AAL92380.1| hypothetical protein [Dictyostelium discoideum]
gi|28829534|gb|AAM33726.2| hypothetical protein
[Dictyostelium discoideum]
Length = 1097
Score = 31.2 bits (69), Expect = 6.6
Identities = 14/39 (35%), Positives = 22/39 (55%)
Frame = -3
Query: 199 PSDLNIVSRCGDSLYNYCISFGYCISISNHRRLDSMNLG 83
PS +N++ C ++ YN C GYC+S + DS + G
Sbjct: 604 PSGINVILNCSEN-YNDCNGNGYCLSDIGQCKCDSNHQG 641
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 416,267,713
Number of Sequences: 1393205
Number of extensions: 8527040
Number of successful extensions: 19853
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 19238
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 19833
length of database: 448,689,247
effective HSP length: 114
effective length of database: 289,863,877
effective search space used: 14493193850
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)