Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC010138A_C01 KMC010138A_c01
(469 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
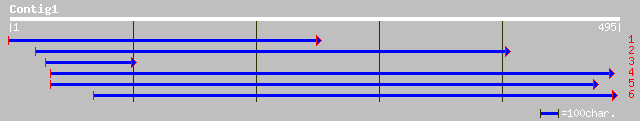
ref|NP_567628.1| putative protein; protein id: At4g21460.1, supp... 44 0.001
pir||T05154 hypothetical protein F18E5.80 - Arabidopsis thaliana... 44 0.001
ref|NP_566603.1| expressed protein; protein id: At3g18240.1, sup... 44 0.001
ref|NP_737044.1| putative phosphoserine phosphatase [Corynebacte... 30 9.5
>ref|NP_567628.1| putative protein; protein id: At4g21460.1, supported by cDNA:
10221. [Arabidopsis thaliana]
Length = 415
Score = 43.5 bits (101), Expect = 0.001
Identities = 21/34 (61%), Positives = 26/34 (75%)
Frame = -2
Query: 462 RPSYAKERLRANPAFMARLHAKSMRVRESNQIPA 361
R SY K+RLRA+PAFM +L AK +R +ESN I A
Sbjct: 382 RTSYVKQRLRASPAFMQKLQAKIIRSKESNAINA 415
>pir||T05154 hypothetical protein F18E5.80 - Arabidopsis thaliana
gi|3080390|emb|CAA18710.1| putative protein [Arabidopsis
thaliana] gi|7268943|emb|CAB81253.1| putative protein
[Arabidopsis thaliana]
Length = 420
Score = 43.5 bits (101), Expect = 0.001
Identities = 21/34 (61%), Positives = 26/34 (75%)
Frame = -2
Query: 462 RPSYAKERLRANPAFMARLHAKSMRVRESNQIPA 361
R SY K+RLRA+PAFM +L AK +R +ESN I A
Sbjct: 387 RTSYVKQRLRASPAFMQKLQAKIIRSKESNAINA 420
>ref|NP_566603.1| expressed protein; protein id: At3g18240.1, supported by cDNA:
gi_11908119, supported by cDNA: gi_13194809, supported
by cDNA: gi_13265398 [Arabidopsis thaliana]
gi|9279658|dbj|BAB01174.1|
emb|CAA18710.1~gene_id:MIE15.3~strong similarity to
unknown protein [Arabidopsis thaliana]
gi|11692818|gb|AAG40012.1|AF324661_1 AT3g18240
[Arabidopsis thaliana]
gi|11908120|gb|AAG41489.1|AF326907_1 unknown protein
[Arabidopsis thaliana]
gi|13194810|gb|AAK15567.1|AF349520_1 unknown protein
[Arabidopsis thaliana] gi|21536541|gb|AAM60873.1|
unknown [Arabidopsis thaliana]
Length = 419
Score = 43.5 bits (101), Expect = 0.001
Identities = 21/34 (61%), Positives = 26/34 (75%)
Frame = -2
Query: 462 RPSYAKERLRANPAFMARLHAKSMRVRESNQIPA 361
R SY K+RLRANPAFM +L AK +R +ES+ I A
Sbjct: 386 RTSYVKQRLRANPAFMQKLQAKIIRSKESDAINA 419
>ref|NP_737044.1| putative phosphoserine phosphatase [Corynebacterium efficiens
YS-314] gi|23492270|dbj|BAC17244.1| putative
phosphoserine phosphatase [Corynebacterium efficiens
YS-314]
Length = 395
Score = 30.4 bits (67), Expect = 9.5
Identities = 31/112 (27%), Positives = 43/112 (37%)
Frame = +2
Query: 128 NKKDMTKGRPSGVSYHCGRGSDVRSLTPLCSEVVSATMTHDPTQYKSQQFIKETKSGSNN 307
N D+ +GR + + RG V L LC E+V TM D T ++Q +
Sbjct: 192 NAHDVAQGREQALEFI--RGRSVEELVELCEEIVDRTM-FDKTWPGTRQLADMHLAAG-- 246
Query: 308 KLRQVNVPTFIHQTWKP*AGI*LDSRTLMDLACSLAMKAGFARSLSLAYEGR 463
HQ W L S T + LA LA + GF ++ E R
Sbjct: 247 -----------HQVW-------LVSATPVQLAQILAQRFGFTGAIGTVPEVR 280
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 378,222,031
Number of Sequences: 1393205
Number of extensions: 7217586
Number of successful extensions: 12112
Number of sequences better than 10.0: 8
Number of HSP's better than 10.0 without gapping: 11969
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 12110
length of database: 448,689,247
effective HSP length: 113
effective length of database: 291,257,082
effective search space used: 12232797444
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)