Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
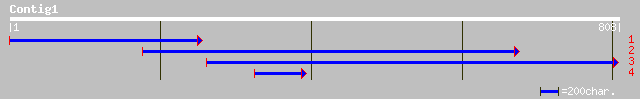
Query= KMC010105A_C01 KMC010105A_c01
(716 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_197617.1| CCR4-associated factor-like protein; protein id... 203 2e-51
ref|NP_190012.1| CCR4-associated factor 1-like protein; protein ... 194 1e-48
ref|NP_565735.1| putative CCR4-associated factor; protein id: At... 158 8e-38
pir||F84728 probable CCR4-associated factor [imported] - Arabido... 158 8e-38
ref|NP_196657.1| CCR4-ASSOCIATED FACTOR -like protein; protein ... 153 3e-36
>ref|NP_197617.1| CCR4-associated factor-like protein; protein id: At5g22250.1,
supported by cDNA: gi_17381057 [Arabidopsis thaliana]
gi|9757805|dbj|BAB08323.1| CCR4-associated factor-like
protein [Arabidopsis thaliana]
gi|17381058|gb|AAL36341.1| putative CCR4-associated
factor [Arabidopsis thaliana] gi|25054979|gb|AAN71961.1|
putative CCR4-associated factor [Arabidopsis thaliana]
Length = 278
Score = 203 bits (517), Expect = 2e-51
Identities = 101/144 (70%), Positives = 120/144 (83%), Gaps = 1/144 (0%)
Frame = -2
Query: 715 RNREFGIDSFRFAELMMSSGLVCDDAVSWVTFHSAYDFGYLVKLLTQRALPEELMEFLRL 536
RNR G++S RFAELMMSSGL+C+++VSWVTFHSAYDFGYLVK+LT+R LP L EFL L
Sbjct: 136 RNRREGVESERFAELMMSSGLICNESVSWVTFHSAYDFGYLVKILTRRQLPVALREFLGL 195
Query: 535 VRVFFGDRVFDVKHLMRFC-SNLHGGLDRVCHSLKVERVIGKSHQAGSDSLLTLHAFQNI 359
+R FFGDRV+DVKH+MRFC L+GGLDRV SL+V R +GK HQAGSDSLLT AFQ +
Sbjct: 196 LRAFFGDRVYDVKHIMRFCEQRLYGGLDRVARSLEVNRAVGKCHQAGSDSLLTWQAFQRM 255
Query: 358 REVYFGKADGLVKYAGVLYGLEVF 287
R++YF + DG K+AGVLYGLEVF
Sbjct: 256 RDLYFVE-DGAEKHAGVLYGLEVF 278
>ref|NP_190012.1| CCR4-associated factor 1-like protein; protein id: At3g44260.1
[Arabidopsis thaliana] gi|11288396|pir||T49142
CCR4-associated factor 1-like protein - Arabidopsis
thaliana gi|7649377|emb|CAB88994.1| CCR4-associated
factor 1-like protein [Arabidopsis thaliana]
gi|15292829|gb|AAK92783.1| putative CCR4-associated
factor 1 [Arabidopsis thaliana]
gi|21436313|gb|AAM51295.1| putative CCR4-associated
factor 1 [Arabidopsis thaliana]
Length = 280
Score = 194 bits (492), Expect = 1e-48
Identities = 97/144 (67%), Positives = 118/144 (81%), Gaps = 1/144 (0%)
Frame = -2
Query: 715 RNREFGIDSFRFAELMMSSGLVCDDAVSWVTFHSAYDFGYLVKLLTQRALPEELMEFLRL 536
RN G++S RFAELMMSSGLVC++ VSWVTFHSAYDFGYL+K+LT+R LP L EF R+
Sbjct: 138 RNCRDGVESERFAELMMSSGLVCNEEVSWVTFHSAYDFGYLMKILTRRELPGALGEFKRV 197
Query: 535 VRVFFGDRVFDVKHLMRFCS-NLHGGLDRVCHSLKVERVIGKSHQAGSDSLLTLHAFQNI 359
+RV FG+RV+DVKH+M+FC L GGLDRV +L+V R +GK HQAGSDSLLT HAFQ +
Sbjct: 198 MRVLFGERVYDVKHMMKFCERRLFGGLDRVARTLEVNRAVGKCHQAGSDSLLTWHAFQRM 257
Query: 358 REVYFGKADGLVKYAGVLYGLEVF 287
R++YF + DG K+AGVLYGLEVF
Sbjct: 258 RDLYFVQ-DGPEKHAGVLYGLEVF 280
>ref|NP_565735.1| putative CCR4-associated factor; protein id: At2g32070.1, supported
by cDNA: gi_15293024 [Arabidopsis thaliana]
gi|15293025|gb|AAK93623.1| putative CCR4-associated
factor [Arabidopsis thaliana] gi|20197620|gb|AAD15397.2|
putative CCR4-associated factor [Arabidopsis thaliana]
gi|23296713|gb|AAN13153.1| putative CCR4-associated
factor [Arabidopsis thaliana]
Length = 275
Score = 158 bits (399), Expect = 8e-38
Identities = 82/142 (57%), Positives = 106/142 (73%)
Frame = -2
Query: 715 RNREFGIDSFRFAELMMSSGLVCDDAVSWVTFHSAYDFGYLVKLLTQRALPEELMEFLRL 536
+N EFGIDS RFAEL+MSSG+V ++ V WVTFHS YDFGYL+KLLT + LPE F +
Sbjct: 131 KNNEFGIDSKRFAELLMSSGIVLNENVHWVTFHSGYDFGYLLKLLTCQNLPETQTGFFEM 190
Query: 535 VRVFFGDRVFDVKHLMRFCSNLHGGLDRVCHSLKVERVIGKSHQAGSDSLLTLHAFQNIR 356
+ V+F RV+D+KHLM+FC++LHGGL+++ L VERV G HQAGSDSLLT F+ ++
Sbjct: 191 ISVYF-PRVYDIKHLMKFCNSLHGGLNKLAELLDVERV-GICHQAGSDSLLTSCTFRKLQ 248
Query: 355 EVYFGKADGLVKYAGVLYGLEV 290
E +F + KY+GVLYGL V
Sbjct: 249 ENFF--IGSMEKYSGVLYGLGV 268
>pir||F84728 probable CCR4-associated factor [imported] - Arabidopsis thaliana
Length = 252
Score = 158 bits (399), Expect = 8e-38
Identities = 82/142 (57%), Positives = 106/142 (73%)
Frame = -2
Query: 715 RNREFGIDSFRFAELMMSSGLVCDDAVSWVTFHSAYDFGYLVKLLTQRALPEELMEFLRL 536
+N EFGIDS RFAEL+MSSG+V ++ V WVTFHS YDFGYL+KLLT + LPE F +
Sbjct: 108 KNNEFGIDSKRFAELLMSSGIVLNENVHWVTFHSGYDFGYLLKLLTCQNLPETQTGFFEM 167
Query: 535 VRVFFGDRVFDVKHLMRFCSNLHGGLDRVCHSLKVERVIGKSHQAGSDSLLTLHAFQNIR 356
+ V+F RV+D+KHLM+FC++LHGGL+++ L VERV G HQAGSDSLLT F+ ++
Sbjct: 168 ISVYF-PRVYDIKHLMKFCNSLHGGLNKLAELLDVERV-GICHQAGSDSLLTSCTFRKLQ 225
Query: 355 EVYFGKADGLVKYAGVLYGLEV 290
E +F + KY+GVLYGL V
Sbjct: 226 ENFF--IGSMEKYSGVLYGLGV 245
>ref|NP_196657.1| CCR4-ASSOCIATED FACTOR -like protein; protein id: At5g10960.1,
supported by cDNA: gi_15292842 [Arabidopsis thaliana]
gi|11288398|pir||T50805 CCR4-ASSOCIATED FACTOR-like
protein - Arabidopsis thaliana
gi|8979730|emb|CAB96851.1| CCR4-ASSOCIATED FACTOR-like
protein [Arabidopsis thaliana]
gi|23296319|gb|AAN13040.1| putative CCR4-associated
factor [Arabidopsis thaliana]
Length = 277
Score = 153 bits (386), Expect = 3e-36
Identities = 82/142 (57%), Positives = 102/142 (71%)
Frame = -2
Query: 715 RNREFGIDSFRFAELMMSSGLVCDDAVSWVTFHSAYDFGYLVKLLTQRALPEELMEFLRL 536
+N E GID RF ELMMSSG+V +DA+SWVTFH YDFGYLVKLLT + LP + +F +L
Sbjct: 130 KNIEKGIDVVRFGELMMSSGIVLNDAISWVTFHGGYDFGYLVKLLTCKELPLKQADFFKL 189
Query: 535 VRVFFGDRVFDVKHLMRFCSNLHGGLDRVCHSLKVERVIGKSHQAGSDSLLTLHAFQNIR 356
+ V+F V+D+KHLM FC+ L GGL+R+ + VERV G HQAGSDSLLTL +F+ ++
Sbjct: 190 LYVYF-PTVYDIKHLMTFCNGLFGGLNRLAELMGVERV-GICHQAGSDSLLTLGSFRKLK 247
Query: 355 EVYFGKADGLVKYAGVLYGLEV 290
E YF KY GVLYGL V
Sbjct: 248 ERYF--PGSTEKYTGVLYGLGV 267
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 621,201,208
Number of Sequences: 1393205
Number of extensions: 13804850
Number of successful extensions: 40215
Number of sequences better than 10.0: 70
Number of HSP's better than 10.0 without gapping: 37595
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 39981
length of database: 448,689,247
effective HSP length: 120
effective length of database: 281,504,647
effective search space used: 33217548346
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)