Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC010019A_C01 KMC010019A_c01
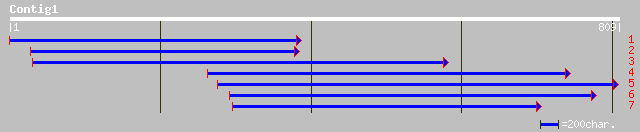
(809 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_568356.1| chloroplast GrpE protein; protein id: At5g17710... 160 2e-38
emb|CAB40381.1| GrpE protein [Arabidopsis thaliana] gi|9759048|d... 160 2e-38
gb|AAM65596.1| putative heat shock protein [Arabidopsis thaliana] 111 1e-23
ref|NP_564475.1| putative heat shock protein; protein id: At1g36... 111 1e-23
gb|AAK96224.1|AF406936_1 co-chaperone CGE1 precursor isoform b [... 110 2e-23
>ref|NP_568356.1| chloroplast GrpE protein; protein id: At5g17710.1, supported by
cDNA: gi_13878046, supported by cDNA: gi_17104678
[Arabidopsis thaliana]
gi|13878047|gb|AAK44101.1|AF370286_1 putative
chloroplast GrpE protein [Arabidopsis thaliana]
gi|17104679|gb|AAL34228.1| putative chloroplast GrpE
protein [Arabidopsis thaliana]
Length = 324
Score = 160 bits (406), Expect = 2e-38
Identities = 83/117 (70%), Positives = 98/117 (82%), Gaps = 1/117 (0%)
Frame = -3
Query: 807 QIKVETEAEEKISNSYQSIYKQFIEILTSLGVEPVETVGKFFDPLLHEAIMREDSTEFEE 628
QIKVETE EEK++NSYQSIYKQF+EIL SLGV VETVGK FDP+LHEAIMREDS E+EE
Sbjct: 210 QIKVETEGEEKVTNSYQSIYKQFVEILGSLGVIHVETVGKQFDPMLHEAIMREDSAEYEE 269
Query: 627 GIIIQEFRKGFQLGDRLLRPSMVKVSAGPGPAKP-ESEAPQEEQVTSEISKENEGGT 460
GI+++E+RKGF LG+RLLRPSMVKVSAGPGP KP E+E E+ T++ S E E +
Sbjct: 270 GIVLEEYRKGFLLGERLLRPSMVKVSAGPGPEKPLEAEG---EEATAQGSAEEESSS 323
>emb|CAB40381.1| GrpE protein [Arabidopsis thaliana] gi|9759048|dbj|BAB09570.1| GrpE
protein [Arabidopsis thaliana]
Length = 326
Score = 160 bits (406), Expect = 2e-38
Identities = 83/117 (70%), Positives = 98/117 (82%), Gaps = 1/117 (0%)
Frame = -3
Query: 807 QIKVETEAEEKISNSYQSIYKQFIEILTSLGVEPVETVGKFFDPLLHEAIMREDSTEFEE 628
QIKVETE EEK++NSYQSIYKQF+EIL SLGV VETVGK FDP+LHEAIMREDS E+EE
Sbjct: 212 QIKVETEGEEKVTNSYQSIYKQFVEILGSLGVIHVETVGKQFDPMLHEAIMREDSAEYEE 271
Query: 627 GIIIQEFRKGFQLGDRLLRPSMVKVSAGPGPAKP-ESEAPQEEQVTSEISKENEGGT 460
GI+++E+RKGF LG+RLLRPSMVKVSAGPGP KP E+E E+ T++ S E E +
Sbjct: 272 GIVLEEYRKGFLLGERLLRPSMVKVSAGPGPEKPLEAEG---EEATAQGSAEEESSS 325
>gb|AAM65596.1| putative heat shock protein [Arabidopsis thaliana]
Length = 279
Score = 111 bits (278), Expect = 1e-23
Identities = 56/100 (56%), Positives = 71/100 (71%)
Frame = -3
Query: 807 QIKVETEAEEKISNSYQSIYKQFIEILTSLGVEPVETVGKFFDPLLHEAIMREDSTEFEE 628
Q++V+T+ E+KI SYQ IY+QF+E+L L V + TVGK FDPLLHEAI RE+S +
Sbjct: 175 QVRVDTDKEKKIDTSYQGIYRQFVEVLRYLRVSVIATVGKPFDPLLHEAISREESEAVKA 234
Query: 627 GIIIQEFRKGFQLGDRLLRPSMVKVSAGPGPAKPESEAPQ 508
GII +E KGF LGDR+LRP+ VKVS GP K S A +
Sbjct: 235 GIITEELNKGFVLGDRVLRPAKVKVSLGPVNKKTPSAAEE 274
>ref|NP_564475.1| putative heat shock protein; protein id: At1g36390.1, supported by
cDNA: 40701., supported by cDNA: gi_17529221
[Arabidopsis thaliana] gi|25403240|pir||H86484 probable
heat shock protein, 54606-52893 [imported] - Arabidopsis
thaliana gi|12324480|gb|AAG52200.1|AC021199_6 putative
heat shock protein; 54606-52893 [Arabidopsis thaliana]
gi|17529222|gb|AAL38838.1| putative heat shock protein
[Arabidopsis thaliana] gi|21436225|gb|AAM51251.1|
putative heat shock protein [Arabidopsis thaliana]
Length = 279
Score = 111 bits (278), Expect = 1e-23
Identities = 56/100 (56%), Positives = 71/100 (71%)
Frame = -3
Query: 807 QIKVETEAEEKISNSYQSIYKQFIEILTSLGVEPVETVGKFFDPLLHEAIMREDSTEFEE 628
Q++V+T+ E+KI SYQ IY+QF+E+L L V + TVGK FDPLLHEAI RE+S +
Sbjct: 175 QVRVDTDKEKKIDTSYQGIYRQFVEVLRYLRVSVIATVGKPFDPLLHEAISREESEAVKA 234
Query: 627 GIIIQEFRKGFQLGDRLLRPSMVKVSAGPGPAKPESEAPQ 508
GII +E KGF LGDR+LRP+ VKVS GP K S A +
Sbjct: 235 GIITEELNKGFVLGDRVLRPAKVKVSLGPVNKKTPSAAEE 274
>gb|AAK96224.1|AF406936_1 co-chaperone CGE1 precursor isoform b [Chlamydomonas reinhardtii]
Length = 260
Score = 110 bits (276), Expect = 2e-23
Identities = 53/98 (54%), Positives = 71/98 (72%), Gaps = 1/98 (1%)
Frame = -3
Query: 807 QIKVETEAEEKISNSYQSIYKQFIEILTSLGVEPVETVGKFFDPLLHEAIMREDSTEFEE 628
Q+K ETEAE+KI+NSYQ +YKQ ++++ + GVE V T G FDP +H+AIMRE S +
Sbjct: 163 QVKAETEAEQKINNSYQGLYKQMVDLMRTQGVEAVPTTGTPFDPNIHDAIMREPSNSHPD 222
Query: 627 GIIIQEFRKGFQLGDRLLRPSMVKVS-AGPGPAKPESE 517
G ++QEFRKGF +G +L+RP+MVKVS GPA E
Sbjct: 223 GTVLQEFRKGFAIGGKLIRPAMVKVSYTEDGPAASSEE 260
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 670,936,325
Number of Sequences: 1393205
Number of extensions: 14355921
Number of successful extensions: 81136
Number of sequences better than 10.0: 216
Number of HSP's better than 10.0 without gapping: 41849
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 63317
length of database: 448,689,247
effective HSP length: 121
effective length of database: 280,111,442
effective search space used: 41456493416
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)