Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC009543A_C01 KMC009543A_c01
(582 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
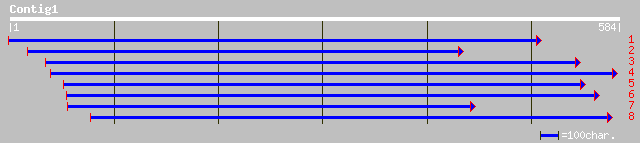
sp|P29502|TBB3_PEA TUBULIN BETA-3 CHAIN gi|81953|pir||S20870 tub... 155 3e-37
dbj|BAA82639.1| Beta-tubulin [Zinnia elegans] 146 2e-34
gb|AAA20243.1| beta-tubulin 146 2e-34
dbj|BAA11392.1| putative tubulin [Brassica rapa] 144 6e-34
dbj|BAB39951.1| putative tubulin beta-4 chain [Oryza sativa (jap... 144 1e-33
>sp|P29502|TBB3_PEA TUBULIN BETA-3 CHAIN gi|81953|pir||S20870 tubulin beta-3 chain -
garden pea (fragment) gi|388255|emb|CAA38615.1|
beta-tubulin 3 [Pisum sativum]
Length = 440
Score = 155 bits (392), Expect = 3e-37
Identities = 73/77 (94%), Positives = 76/77 (97%)
Frame = -3
Query: 580 SIQEMFRRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVSEYQQYQDATAED 401
SIQEMFRRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVSEYQQYQDATA++
Sbjct: 364 SIQEMFRRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVSEYQQYQDATADE 423
Query: 400 DGEYEDEEEEEPEHVYE 350
+GEYEDEEEEEPEH YE
Sbjct: 424 EGEYEDEEEEEPEHGYE 440
>dbj|BAA82639.1| Beta-tubulin [Zinnia elegans]
Length = 441
Score = 146 bits (369), Expect = 2e-34
Identities = 69/74 (93%), Positives = 73/74 (98%)
Frame = -3
Query: 580 SIQEMFRRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVSEYQQYQDATAED 401
SIQEMFRRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVSEYQQYQDATA+D
Sbjct: 366 SIQEMFRRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVSEYQQYQDATADD 425
Query: 400 DGEYEDEEEEEPEH 359
DGEYE+EEEEE ++
Sbjct: 426 DGEYEEEEEEEGDY 439
>gb|AAA20243.1| beta-tubulin
Length = 322
Score = 146 bits (368), Expect = 2e-34
Identities = 69/73 (94%), Positives = 72/73 (98%)
Frame = -3
Query: 580 SIQEMFRRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVSEYQQYQDATAED 401
SIQEMFRRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVSEYQQYQDATAED
Sbjct: 246 SIQEMFRRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVSEYQQYQDATAED 305
Query: 400 DGEYEDEEEEEPE 362
DGEYEDEE+++ E
Sbjct: 306 DGEYEDEEDDDVE 318
>dbj|BAA11392.1| putative tubulin [Brassica rapa]
Length = 88
Score = 144 bits (364), Expect = 6e-34
Identities = 68/74 (91%), Positives = 73/74 (97%)
Frame = -3
Query: 580 SIQEMFRRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVSEYQQYQDATAED 401
SIQEMFRRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVSEYQQYQDATA++
Sbjct: 13 SIQEMFRRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVSEYQQYQDATADE 72
Query: 400 DGEYEDEEEEEPEH 359
+GEYE+EEEEE E+
Sbjct: 73 EGEYEEEEEEEVEY 86
>dbj|BAB39951.1| putative tubulin beta-4 chain [Oryza sativa (japonica
cultivar-group)] gi|15408819|dbj|BAB64211.1| tubulin
beta-4 chain [Oryza sativa (japonica cultivar-group)]
Length = 447
Score = 144 bits (362), Expect = 1e-33
Identities = 67/73 (91%), Positives = 72/73 (97%)
Frame = -3
Query: 580 SIQEMFRRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVSEYQQYQDATAED 401
SIQEMFRRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVSEYQQYQDATA++
Sbjct: 373 SIQEMFRRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVSEYQQYQDATADE 432
Query: 400 DGEYEDEEEEEPE 362
+G+YEDEEE+ PE
Sbjct: 433 EGDYEDEEEQVPE 445
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 496,671,920
Number of Sequences: 1393205
Number of extensions: 10681817
Number of successful extensions: 93365
Number of sequences better than 10.0: 1209
Number of HSP's better than 10.0 without gapping: 55196
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 76704
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 21712003912
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)