Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
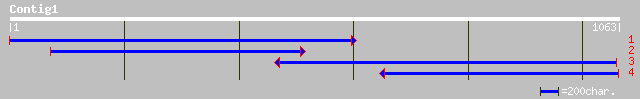
Query= KMC009434A_C01 KMC009434A_c01
(1058 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_200279.1| bHLH protein; protein id: At5g54680.1, supporte... 330 3e-89
gb|AAM64276.1| bHLH transcription factor, putative [Arabidopsis ... 328 9e-89
ref|NP_175518.1| bHLH protein; protein id: At1g51070.1, supporte... 277 2e-73
gb|AAO72577.1| helix-loop-helix-like protein [Oryza sativa (japo... 228 9e-59
gb|AAM10939.1|AF488573_1 putative bHLH transcription factor [Ara... 184 3e-45
>ref|NP_200279.1| bHLH protein; protein id: At5g54680.1, supported by cDNA: 22343.,
supported by cDNA: gi_15451009, supported by cDNA:
gi_20127110 [Arabidopsis thaliana]
gi|10176795|dbj|BAB09934.1|
gene_id:K5F14.2~pir||B71406~similar to unknown protein
[Arabidopsis thaliana] gi|15451010|gb|AAK96776.1|
Unknown protein [Arabidopsis thaliana]
gi|20127111|gb|AAM10964.1|AF488629_1 putative bHLH
transcription factor [Arabidopsis thaliana]
gi|25084222|gb|AAN72200.1| Unknown protein [Arabidopsis
thaliana]
Length = 234
Score = 330 bits (845), Expect = 3e-89
Identities = 174/240 (72%), Positives = 197/240 (81%), Gaps = 2/240 (0%)
Frame = +3
Query: 189 MVSRENTNWLFDYGLIDDIPAPEVTFTVPPSGFTWPSSQPLNSSSNVVGVEIDGSLGDSD 368
MVS EN NW+ D LID A +FT+ GF+WP QP+ SSN +DGS G+S+
Sbjct: 1 MVSPENANWICD--LID---ADYGSFTIQGPGFSWPVQQPIGVSSNS-SAGVDGSAGNSE 54
Query: 369 SLKEPGSKKRVRSESCSATSSKACREKLRRDKLNDKFVELGSILEPGRPPQTDKAAILID 548
+ KEPGSKKR R ES SATSSKACREK RRD+LNDKF+ELG+ILEPG PP+TDKAAIL+D
Sbjct: 55 ASKEPGSKKRGRCESSSATSSKACREKQRRDRLNDKFMELGAILEPGNPPKTDKAAILVD 114
Query: 549 AVRMVTQLRGEAQKMKDTNMGLQEKIKELKTEKNELRDEKQRLKTEKERLEQQLKSMNA- 725
AVRMVTQLRGEAQK+KD+N LQ+KIKELKTEKNELRDEKQRLKTEKE+LEQQLK+MNA
Sbjct: 115 AVRMVTQLRGEAQKLKDSNSSLQDKIKELKTEKNELRDEKQRLKTEKEKLEQQLKAMNAP 174
Query: 726 QPSFMPPPQALPAAFA-AQGQAHGNKLVPFISYPGVAMWQFLPPAARDTSQDHELRPPVA 902
QPSF P P +P AFA AQGQA GNK+VP ISYPGVAMWQF+PPA+ DTSQDH LRPPVA
Sbjct: 175 QPSFFPAPPMMPTAFASAQGQAPGNKMVPIISYPGVAMWQFMPPASVDTSQDHVLRPPVA 234
>gb|AAM64276.1| bHLH transcription factor, putative [Arabidopsis thaliana]
Length = 234
Score = 328 bits (841), Expect = 9e-89
Identities = 173/240 (72%), Positives = 197/240 (82%), Gaps = 2/240 (0%)
Frame = +3
Query: 189 MVSRENTNWLFDYGLIDDIPAPEVTFTVPPSGFTWPSSQPLNSSSNVVGVEIDGSLGDSD 368
MVS EN NW+ D LID A +FT+ GF+WP QP+ SSN +DGS G+S+
Sbjct: 1 MVSPENANWICD--LID---ADYGSFTIQGPGFSWPVQQPIGVSSNS-SAGVDGSAGNSE 54
Query: 369 SLKEPGSKKRVRSESCSATSSKACREKLRRDKLNDKFVELGSILEPGRPPQTDKAAILID 548
+ KEPGSKKR R ES SATSSKACREK RRD+LNDKF+ELG+ILEPG PP+TDKAAIL+D
Sbjct: 55 ASKEPGSKKRGRCESSSATSSKACREKQRRDRLNDKFMELGAILEPGNPPKTDKAAILVD 114
Query: 549 AVRMVTQLRGEAQKMKDTNMGLQEKIKELKTEKNELRDEKQRLKTEKERLEQQLKSMNA- 725
AVRMVTQLRGEAQK+KD+N LQ+KIKELKTEKNELRDEKQRLKTEKE+LEQQLK++NA
Sbjct: 115 AVRMVTQLRGEAQKLKDSNSSLQDKIKELKTEKNELRDEKQRLKTEKEKLEQQLKAINAP 174
Query: 726 QPSFMPPPQALPAAFA-AQGQAHGNKLVPFISYPGVAMWQFLPPAARDTSQDHELRPPVA 902
QPSF P P +P AFA AQGQA GNK+VP ISYPGVAMWQF+PPA+ DTSQDH LRPPVA
Sbjct: 175 QPSFFPAPPMMPTAFASAQGQAPGNKMVPIISYPGVAMWQFMPPASVDTSQDHVLRPPVA 234
>ref|NP_175518.1| bHLH protein; protein id: At1g51070.1, supported by cDNA: 16284.,
supported by cDNA: gi_20127114 [Arabidopsis thaliana]
gi|25405430|pir||H96547 probable bHLH transcription
factor [imported] - Arabidopsis thaliana
gi|12320783|gb|AAG50538.1|AC079828_9 bHLH transcription
factor, putative [Arabidopsis thaliana]
gi|20127115|gb|AAM10965.1|AF488632_1 putative bHLH
transcription factor [Arabidopsis thaliana]
gi|21553747|gb|AAM62840.1| bHLH transcription factor,
putative [Arabidopsis thaliana]
gi|27311655|gb|AAO00793.1| bHLH transcription factor,
putative [Arabidopsis thaliana]
Length = 226
Score = 277 bits (709), Expect = 2e-73
Identities = 147/242 (60%), Positives = 174/242 (71%), Gaps = 4/242 (1%)
Frame = +3
Query: 189 MVSRENTNWLFDYGLIDDIPAPEVTFTVPPSGFTWPSSQPLNSSSNVVGVEIDGSLGDSD 368
MVS ENTNWL DY LI E F+ F W S V VE+DG L D+D
Sbjct: 1 MVSPENTNWLSDYPLI------EGAFSDQNPTFPWQID-----GSATVSVEVDGFLCDAD 49
Query: 369 SLKEPGSKKRVRSESCSATSSKACREKLRRDKLNDKFVELGSILEPGRPPQTDKAAILID 548
+KEP S+KR+++ESC+ ++SKACREK RRD+LNDKF EL S+LEPGR P+TDK AI+ D
Sbjct: 50 VIKEPSSRKRIKTESCTGSNSKACREKQRRDRLNDKFTELSSVLEPGRTPKTDKVAIIND 109
Query: 549 AVRMVTQLRGEAQKMKDTNMGLQEKIKELKTEKNELRDEKQRLKTEKERLEQQLKSMNAQ 728
A+RMV Q R EAQK+KD N LQEKIKELK EKNELRDEKQ+LK EKER++QQLK++ Q
Sbjct: 110 AIRMVNQARDEAQKLKDLNSSLQEKIKELKDEKNELRDEKQKLKVEKERIDQQLKAIKTQ 169
Query: 729 PS----FMPPPQALPAAFAAQGQAHGNKLVPFISYPGVAMWQFLPPAARDTSQDHELRPP 896
P F+P PQ L +Q QA G+KLVPF +YPG AMWQF+PPAA DTSQDH LRPP
Sbjct: 170 PQPQPCFLPNPQTL-----SQAQAPGSKLVPFTTYPGFAMWQFMPPAAVDTSQDHVLRPP 224
Query: 897 VA 902
VA
Sbjct: 225 VA 226
>gb|AAO72577.1| helix-loop-helix-like protein [Oryza sativa (japonica
cultivar-group)]
Length = 216
Score = 228 bits (582), Expect = 9e-59
Identities = 121/216 (56%), Positives = 154/216 (71%), Gaps = 10/216 (4%)
Frame = +3
Query: 285 FTWPSSQPLNSSSNVVGVEIDGSLGDSDSLKEPGSKKRVRSESCSATSSKACREKLRRDK 464
F W +S ++ S V ++ + S LKEPGS KRVRS SC +SKA REK+RRDK
Sbjct: 4 FPWDASPSCSNPSVEVSSYVNTT---SYVLKEPGSNKRVRSGSCGRPTSKASREKIRRDK 60
Query: 465 LNDKFVELGSILEPGRPPQTDKAAILIDAVRMVTQLRGEAQKMKDTNMGLQEKIKELKTE 644
+ND+F+ELG+ LEPG+P ++DKAAIL DA RMV QLR EA+++KDTN L++KIKELK E
Sbjct: 61 MNDRFLELGTTLEPGKPVKSDKAAILSDATRMVIQLRAEAKQLKDTNESLEDKIKELKAE 120
Query: 645 KNELRDEKQRLKTEKERLEQQLKSMNAQPSFMPPPQALPAAFA---------AQGQAHGN 797
K+ELRDEKQ+LK EKE LEQQ+K + A P++MP P +PA + AQGQA G
Sbjct: 121 KDELRDEKQKLKVEKETLEQQVKILTATPAYMPHPTLMPAPYPQAPLAPFHHAQGQAAGQ 180
Query: 798 KL-VPFISYPGVAMWQFLPPAARDTSQDHELRPPVA 902
KL +PF+ YPG MWQF+PP+ DTS+D E PPVA
Sbjct: 181 KLMMPFVGYPGYPMWQFMPPSEVDTSKDSEACPPVA 216
>gb|AAM10939.1|AF488573_1 putative bHLH transcription factor [Arabidopsis thaliana]
Length = 291
Score = 184 bits (466), Expect = 3e-45
Identities = 106/200 (53%), Positives = 134/200 (67%), Gaps = 6/200 (3%)
Frame = +3
Query: 321 SNVVGVEIDGSLGD-SDSLKEPGSKKRVRSESCSATSSKACREKLRRDKLNDKFVELGSI 497
S VV + S+G + +E S KR R+ SCS +KACREKLRR+KLNDKF++L S+
Sbjct: 100 SGVVEINSSSSVGAVKEEFEEECSGKRRRTGSCSKPGTKACREKLRREKLNDKFMDLSSV 159
Query: 498 LEPGRPPQTDKAAILIDAVRMVTQLRGEAQKMKDTNMGLQEKIKELKTEKNELRDEKQRL 677
LEPGR P+TDK+AIL DA+R+V QLRGEA ++++TN L E+IK LK +KNELR+EK L
Sbjct: 160 LEPGRTPKTDKSAILDDAIRVVNQLRGEAHELQETNQKLLEEIKSLKADKNELREEKLVL 219
Query: 678 KTEKERLEQQLKSMNA-QPSFMPPPQALPAAFAAQGQAHGNKLVPFISY----PGVAMWQ 842
K EKE++EQQLKSM P FMP PAAF H +K+ Y P + MW
Sbjct: 220 KAEKEKMEQQLKSMVVPSPGFMPSQH--PAAF------HSHKMAVAYPYGYYPPNMPMWS 271
Query: 843 FLPPAARDTSQDHELRPPVA 902
LPPA RDTS+D + PPVA
Sbjct: 272 PLPPADRDTSRDLKNLPPVA 291
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 967,645,749
Number of Sequences: 1393205
Number of extensions: 23120444
Number of successful extensions: 106069
Number of sequences better than 10.0: 815
Number of HSP's better than 10.0 without gapping: 82855
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 103901
length of database: 448,689,247
effective HSP length: 124
effective length of database: 275,931,827
effective search space used: 62912456556
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)