Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC009426A_C01 KMC009426A_c01
(608 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
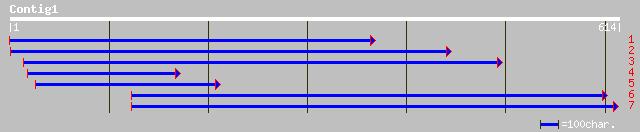
ref|NP_563798.1| expressed protein; protein id: At1g07840.1, sup... 55 7e-07
ref|XP_233232.1| similar to Hypothetical protein DJ1198H6.2 [Rat... 33 2.2
ref|XP_233233.1| similar to Hypothetical protein DJ1198H6.2 [Rat... 33 2.2
ref|NP_500674.1| 7 transmembrane chemoreceptor family member fam... 32 5.0
ref|NP_702466.1| hypothetical protein [Plasmodium falciparum 3D7... 32 8.5
>ref|NP_563798.1| expressed protein; protein id: At1g07840.1, supported by cDNA:
gi_15810572, supported by cDNA: gi_20259572 [Arabidopsis
thaliana] gi|15810573|gb|AAL07174.1| unknown protein
[Arabidopsis thaliana] gi|20259573|gb|AAM14129.1|
unknown protein [Arabidopsis thaliana]
Length = 312
Score = 55.1 bits (131), Expect = 7e-07
Identities = 33/69 (47%), Positives = 44/69 (62%)
Frame = -1
Query: 608 LFNRVPLTKHERKREKYLKKSRNGLDGLTESFFDEIKTLPFEDETGEQVMGPSKGSNRNG 429
LF R P TK ++KREK LK S +GL LTE+F+D+IK F D+ GE+ + + R G
Sbjct: 249 LFTRAPRTKEDKKREKRLKSS-SGLHELTENFYDDIK---FLDKDGEKPRSFGR-NKRGG 303
Query: 428 RLKKRKRKH 402
KKRK +H
Sbjct: 304 PFKKRKTRH 312
>ref|XP_233232.1| similar to Hypothetical protein DJ1198H6.2 [Rattus norvegicus]
Length = 379
Score = 33.5 bits (75), Expect = 2.2
Identities = 13/29 (44%), Positives = 18/29 (61%)
Frame = +3
Query: 81 HIPHVSQNGVHTDHSQKKNIFHWLKSPLQ 167
H+ H+S NG H + K +F LKSPL+
Sbjct: 236 HLQHLSMNGAHFANDSMKELFRCLKSPLE 264
>ref|XP_233233.1| similar to Hypothetical protein DJ1198H6.2 [Rattus norvegicus]
Length = 412
Score = 33.5 bits (75), Expect = 2.2
Identities = 13/29 (44%), Positives = 18/29 (61%)
Frame = +3
Query: 81 HIPHVSQNGVHTDHSQKKNIFHWLKSPLQ 167
H+ H+S NG H + K +F LKSPL+
Sbjct: 269 HLQHLSMNGAHFANDNMKELFRCLKSPLE 297
>ref|NP_500674.1| 7 transmembrane chemoreceptor family member family member
[Caenorhabditis elegans] gi|7332247|gb|AAF60934.1|
Hypothetical protein Y9C9A.10 [Caenorhabditis elegans]
Length = 340
Score = 32.3 bits (72), Expect = 5.0
Identities = 20/57 (35%), Positives = 31/57 (54%), Gaps = 3/57 (5%)
Frame = -1
Query: 218 NVFLLLPCFLL---PLFTVNLEWRLQPMKNVFFLAMISVHPIL*DMWYVDTFHPVLF 57
+VFL LP L+ PLF +N+ W P+ +F +M SV + ++ VD + LF
Sbjct: 261 SVFLFLPTLLIAAGPLFKLNIPWASLPLSILF--SMFSVMDSIPIIFLVDEYRNALF 315
>ref|NP_702466.1| hypothetical protein [Plasmodium falciparum 3D7]
gi|23497650|gb|AAN37190.1|AE014825_49 hypothetical
protein [Plasmodium falciparum 3D7]
Length = 1632
Score = 31.6 bits (70), Expect = 8.5
Identities = 23/75 (30%), Positives = 32/75 (42%), Gaps = 16/75 (21%)
Frame = -1
Query: 584 KHERKREKYLKKSRNGLDGLTESFFDEIKTLPFE----------DETG------EQVMGP 453
K+ ++EK L + L E+F D+ + L E DE G M
Sbjct: 219 KNNIRKEKVLNSGNENMINLNENFLDDNRDLENELKMKAEKCIDDEKGGIYDHNNDNMVK 278
Query: 452 SKGSNRNGRLKKRKR 408
KGSN GR +KRK+
Sbjct: 279 GKGSNAKGRRRKRKK 293
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 558,571,663
Number of Sequences: 1393205
Number of extensions: 12712430
Number of successful extensions: 30326
Number of sequences better than 10.0: 11
Number of HSP's better than 10.0 without gapping: 28701
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 30242
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 24283162270
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)