Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC009241A_C01 KMC009241A_c01
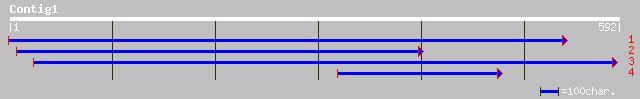
(590 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
emb|CAC87663.1| hyaluronidase [Streptococcus suis] 42 0.006
emb|CAC87664.1| hyaluronidase [Streptococcus suis] 40 0.022
emb|CAC87662.1| hyaluronidase [Streptococcus suis] 40 0.022
ref|NP_703411.1| hypothetical protein [Plasmodium falciparum 3D7... 36 0.42
ref|NP_703568.1| hypothetical protein [Plasmodium falciparum 3D7... 35 0.54
>emb|CAC87663.1| hyaluronidase [Streptococcus suis]
Length = 1164
Score = 42.0 bits (97), Expect = 0.006
Identities = 23/49 (46%), Positives = 27/49 (54%)
Frame = +2
Query: 26 FLYINKYNINYKIDSNWLRYHLFFLLNSHALVNTLQSFFPYFSTSIIIT 172
+LY N YN N ID NW Y + S A+VNTL PYFS I+T
Sbjct: 450 WLYANVYNENKSIDGNWWDYEIG---TSRAVVNTLIYMHPYFSQEEILT 495
>emb|CAC87664.1| hyaluronidase [Streptococcus suis]
Length = 522
Score = 40.0 bits (92), Expect = 0.022
Identities = 22/49 (44%), Positives = 26/49 (52%)
Frame = +2
Query: 26 FLYINKYNINYKIDSNWLRYHLFFLLNSHALVNTLQSFFPYFSTSIIIT 172
+LY N YN N ID NW Y + A+VNTL PYFS I+T
Sbjct: 458 WLYTNVYNENKSIDGNWWDYEIG---TPRAVVNTLIYMHPYFSQEEILT 503
>emb|CAC87662.1| hyaluronidase [Streptococcus suis]
Length = 1164
Score = 40.0 bits (92), Expect = 0.022
Identities = 22/49 (44%), Positives = 26/49 (52%)
Frame = +2
Query: 26 FLYINKYNINYKIDSNWLRYHLFFLLNSHALVNTLQSFFPYFSTSIIIT 172
+LY N YN N ID NW Y + A+VNTL PYFS I+T
Sbjct: 450 WLYTNVYNENKSIDGNWWDYEIG---TPRAVVNTLIYMHPYFSQEEILT 495
>ref|NP_703411.1| hypothetical protein [Plasmodium falciparum 3D7]
gi|23504551|emb|CAD51431.1| hypothetical protein
[Plasmodium falciparum 3D7]
Length = 3008
Score = 35.8 bits (81), Expect = 0.42
Identities = 13/48 (27%), Positives = 27/48 (56%)
Frame = +1
Query: 187 SSDKYSQTKKNKNNNNSRYLYLYHYSLQSTLFFHHNKTSREGQLRNFA 330
+ D + NKN +++ Y+ +Y + +FHH K+ + G+ +NF+
Sbjct: 115 NDDHNNNNNNNKNKSSTSYIIHNNYIDNNNKYFHHTKSKQNGEKKNFS 162
>ref|NP_703568.1| hypothetical protein [Plasmodium falciparum 3D7]
gi|23504710|emb|CAD51588.1| hypothetical protein
[Plasmodium falciparum 3D7]
Length = 9341
Score = 35.4 bits (80), Expect = 0.54
Identities = 35/154 (22%), Positives = 71/154 (45%), Gaps = 7/154 (4%)
Frame = +2
Query: 29 LYINKYNINYK--IDSNWLRYHLFFLLNSHALVNTLQSFFPYFSTSIIITQLN---NKVL 193
LYI+ Y++NYK L H+ N + +L +F+ ++S+++ I N N +L
Sbjct: 2160 LYIDDYHMNYKNMKPQIHLDIHINKWTNEKIIKISLSNFYFFYSSTLFILFSNVVINYIL 2219
Query: 194 TNILKRKKIKIIIIAAIYICTTTLYSLHSSFTTTKLHERGS*GISQHCTPKTASLNRQKC 373
T I +++K+ I+ ++ ++ S +T L +G T N+
Sbjct: 2220 TYIQEKQKLNIL----LFQYDNFRSAISSKYTNPHLENKG----------YTYDKNKSYT 2265
Query: 374 PWDIYLVK--PRHKLVEVELFERFFLLVTRQHNV 469
D Y + R+KL +++L+ F+ + + N+
Sbjct: 2266 SIDKYTLHFLSRNKLKKIKLYNDKFVSIEQIDNI 2299
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 487,075,284
Number of Sequences: 1393205
Number of extensions: 10555460
Number of successful extensions: 33808
Number of sequences better than 10.0: 39
Number of HSP's better than 10.0 without gapping: 28791
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 33248
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 22569056698
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)