Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
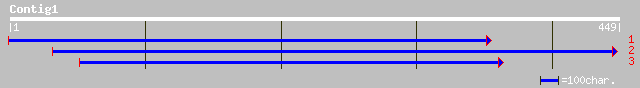
Query= KMC009216A_C01 KMC009216A_c01
(416 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_182041.1| putative SCARECROW gene regulator; protein id: ... 63 2e-13
gb|AAO42084.1| putative SCARECROW gene regulator [Arabidopsis th... 63 2e-13
gb|AAM90848.1|AF481952_1 hairy meristem [Petunia x hybrida] 54 5e-13
ref|NP_191926.1| scarecrow-like 6 (SCL6); protein id: At4g00150.... 57 2e-12
pir||T51237 scarecrow-like protein 6 [imported] - Arabidopsis th... 57 2e-12
>ref|NP_182041.1| putative SCARECROW gene regulator; protein id: At2g45160.1
[Arabidopsis thaliana] gi|25408931|pir||B84887 probable
SCARECROW gene regulator [imported] - Arabidopsis
thaliana
Length = 640
Score = 63.2 bits (152), Expect(3) = 2e-13
Identities = 24/31 (77%), Positives = 30/31 (96%)
Frame = -1
Query: 275 PVRGFHVEKRQSSLVLCWQRKDLISISTWRC 183
PVRGFHVEKRQSSLV+CWQRK+L+++S W+C
Sbjct: 610 PVRGFHVEKRQSSLVMCWQRKELVTVSAWKC 640
Score = 26.2 bits (56), Expect(3) = 2e-13
Identities = 12/31 (38%), Positives = 17/31 (54%)
Frame = -2
Query: 415 EKLVLGRQRLQETTLPWKNMLLSSGFSALPL 323
EKL++ R R E + PW+ + GFS L
Sbjct: 564 EKLLMKRHRWIERSPPWRILFTQCGFSPASL 594
Score = 25.4 bits (54), Expect(3) = 2e-13
Identities = 10/18 (55%), Positives = 12/18 (66%)
Frame = -3
Query: 333 PCHFSNFTESQAECLVQR 280
P S E+QAECL+QR
Sbjct: 591 PASLSQMAEAQAECLLQR 608
>gb|AAO42084.1| putative SCARECROW gene regulator [Arabidopsis thaliana]
Length = 640
Score = 63.2 bits (152), Expect(3) = 2e-13
Identities = 24/31 (77%), Positives = 30/31 (96%)
Frame = -1
Query: 275 PVRGFHVEKRQSSLVLCWQRKDLISISTWRC 183
PVRGFHVEKRQSSLV+CWQRK+L+++S W+C
Sbjct: 610 PVRGFHVEKRQSSLVMCWQRKELVTVSAWKC 640
Score = 26.2 bits (56), Expect(3) = 2e-13
Identities = 12/31 (38%), Positives = 17/31 (54%)
Frame = -2
Query: 415 EKLVLGRQRLQETTLPWKNMLLSSGFSALPL 323
EKL++ R R E + PW+ + GFS L
Sbjct: 564 EKLLMKRHRWIERSPPWRILFTQCGFSPASL 594
Score = 25.4 bits (54), Expect(3) = 2e-13
Identities = 10/18 (55%), Positives = 12/18 (66%)
Frame = -3
Query: 333 PCHFSNFTESQAECLVQR 280
P S E+QAECL+QR
Sbjct: 591 PASLSQMAEAQAECLLQR 608
>gb|AAM90848.1|AF481952_1 hairy meristem [Petunia x hybrida]
Length = 721
Score = 53.9 bits (128), Expect(3) = 5e-13
Identities = 22/30 (73%), Positives = 27/30 (89%)
Frame = -1
Query: 272 VRGFHVEKRQSSLVLCWQRKDLISISTWRC 183
V GFHVEKRQSSLVLCW++++L+S TWRC
Sbjct: 692 VGGFHVEKRQSSLVLCWKQQELLSALTWRC 721
Score = 30.4 bits (67), Expect(3) = 5e-13
Identities = 13/19 (68%), Positives = 15/19 (78%)
Frame = -3
Query: 333 PCHFSNFTESQAECLVQRT 277
P FSN TE QAEC+V+RT
Sbjct: 672 PVAFSNLTEIQAECVVKRT 690
Score = 28.9 bits (63), Expect(3) = 5e-13
Identities = 12/27 (44%), Positives = 17/27 (62%)
Frame = -2
Query: 415 EKLVLGRQRLQETTLPWKNMLLSSGFS 335
E +VLGR R + W+N+ S+GFS
Sbjct: 645 ENMVLGRLRSPDRMPQWRNLFASAGFS 671
>ref|NP_191926.1| scarecrow-like 6 (SCL6); protein id: At4g00150.1, supported by
cDNA: gi_18087576 [Arabidopsis thaliana]
gi|7486541|pir||T01343 hypothetical protein F6N15.20 -
Arabidopsis thaliana gi|3193314|gb|AAC19296.1| contains
similarity to Arabidopsis scarecrow (GB:U62798)
[Arabidopsis thaliana] gi|7267102|emb|CAB80773.1|
scarecrow-like 6 (SCL6) [Arabidopsis thaliana]
gi|18087577|gb|AAL58919.1|AF462831_1 AT4g00150/F6N15_20
[Arabidopsis thaliana] gi|22137044|gb|AAM91367.1|
At4g00150/F6N15_20 [Arabidopsis thaliana]
Length = 558
Score = 57.0 bits (136), Expect(2) = 2e-12
Identities = 21/31 (67%), Positives = 27/31 (86%)
Frame = -1
Query: 275 PVRGFHVEKRQSSLVLCWQRKDLISISTWRC 183
PVRGFHVEK+ +SL+LCWQR +L+ +S WRC
Sbjct: 525 PVRGFHVEKKHNSLLLCWQRTELVGVSAWRC 555
Score = 35.0 bits (79), Expect(2) = 2e-12
Identities = 16/19 (84%), Positives = 16/19 (84%)
Frame = -3
Query: 333 PCHFSNFTESQAECLVQRT 277
P SNFTESQAECLVQRT
Sbjct: 506 PVTHSNFTESQAECLVQRT 524
>pir||T51237 scarecrow-like protein 6 [imported] - Arabidopsis thaliana
(fragment) gi|4580519|gb|AAD24406.1|AF036303_1
scarecrow-like 6 [Arabidopsis thaliana]
Length = 378
Score = 57.0 bits (136), Expect(2) = 2e-12
Identities = 21/31 (67%), Positives = 27/31 (86%)
Frame = -1
Query: 275 PVRGFHVEKRQSSLVLCWQRKDLISISTWRC 183
PVRGFHVEK+ +SL+LCWQR +L+ +S WRC
Sbjct: 345 PVRGFHVEKKHNSLLLCWQRTELVGVSAWRC 375
Score = 35.0 bits (79), Expect(2) = 2e-12
Identities = 16/19 (84%), Positives = 16/19 (84%)
Frame = -3
Query: 333 PCHFSNFTESQAECLVQRT 277
P SNFTESQAECLVQRT
Sbjct: 326 PVTHSNFTESQAECLVQRT 344
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 353,094,825
Number of Sequences: 1393205
Number of extensions: 7351921
Number of successful extensions: 21002
Number of sequences better than 10.0: 34
Number of HSP's better than 10.0 without gapping: 20565
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 20993
length of database: 448,689,247
effective HSP length: 114
effective length of database: 289,863,877
effective search space used: 6956733048
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)