Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC008984A_C01 KMC008984A_c01
(622 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
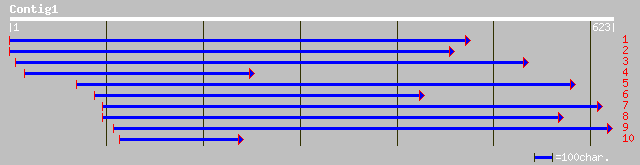
ref|NP_189102.1| chloroplast thylakoidal processing peptidase, p... 187 7e-47
dbj|BAB02011.1| contains similarity to signal peptidase (leader ... 187 7e-47
gb|AAN72018.1| chloroplast thylakoidal processing peptidase, put... 185 4e-46
emb|CAA71502.1| chloroplast thylakoidal processing peptidase [Ar... 150 1e-35
ref|NP_180603.1| putative signal peptidase I; protein id: At2g30... 150 1e-35
>ref|NP_189102.1| chloroplast thylakoidal processing peptidase, putative; protein id:
At3g24590.1 [Arabidopsis thaliana]
Length = 149
Score = 187 bits (476), Expect = 7e-47
Identities = 87/106 (82%), Positives = 94/106 (88%)
Frame = -1
Query: 622 YTDDDVFIKRIVAKEGDIVEVRDGHLIVNGVERNEKFILEPPSYEMKPTRVPENYVFVMG 443
YTD DVFIKRIVAKEGD+VEV +G L+VNGV RNEKFILEPP YEM P RVPEN VFVMG
Sbjct: 42 YTDADVFIKRIVAKEGDLVEVHNGKLMVNGVARNEKFILEPPGYEMTPIRVPENSVFVMG 101
Query: 442 DNRNNSYDSHVWGPLPAKNIIGRSVFRYWPPNRIADTISKEGCAVD 305
DNRNNSYDSHVWGPLP KNIIGRSVFRYWPPNR++ T+ + GCAVD
Sbjct: 102 DNRNNSYDSHVWGPLPLKNIIGRSVFRYWPPNRVSGTVLEGGCAVD 147
>dbj|BAB02011.1| contains similarity to signal peptidase (leader
peptidase)~gene_id:MOB24.17 [Arabidopsis thaliana]
Length = 310
Score = 187 bits (476), Expect = 7e-47
Identities = 87/106 (82%), Positives = 94/106 (88%)
Frame = -1
Query: 622 YTDDDVFIKRIVAKEGDIVEVRDGHLIVNGVERNEKFILEPPSYEMKPTRVPENYVFVMG 443
YTD DVFIKRIVAKEGD+VEV +G L+VNGV RNEKFILEPP YEM P RVPEN VFVMG
Sbjct: 203 YTDADVFIKRIVAKEGDLVEVHNGKLMVNGVARNEKFILEPPGYEMTPIRVPENSVFVMG 262
Query: 442 DNRNNSYDSHVWGPLPAKNIIGRSVFRYWPPNRIADTISKEGCAVD 305
DNRNNSYDSHVWGPLP KNIIGRSVFRYWPPNR++ T+ + GCAVD
Sbjct: 263 DNRNNSYDSHVWGPLPLKNIIGRSVFRYWPPNRVSGTVLEGGCAVD 308
>gb|AAN72018.1| chloroplast thylakoidal processing peptidase, putative [Arabidopsis
thaliana]
Length = 291
Score = 185 bits (470), Expect = 4e-46
Identities = 86/106 (81%), Positives = 93/106 (87%)
Frame = -1
Query: 622 YTDDDVFIKRIVAKEGDIVEVRDGHLIVNGVERNEKFILEPPSYEMKPTRVPENYVFVMG 443
YTD DVFIKRIVAKEGD+VEV +G +VNGV RNEKFILEPP YEM P RVPEN VFVMG
Sbjct: 184 YTDADVFIKRIVAKEGDLVEVHNGKQMVNGVARNEKFILEPPGYEMTPIRVPENSVFVMG 243
Query: 442 DNRNNSYDSHVWGPLPAKNIIGRSVFRYWPPNRIADTISKEGCAVD 305
DNRNNSYDSHVWGPLP KNIIGRSVFRYWPPNR++ T+ + GCAVD
Sbjct: 244 DNRNNSYDSHVWGPLPLKNIIGRSVFRYWPPNRVSGTVLEGGCAVD 289
>emb|CAA71502.1| chloroplast thylakoidal processing peptidase [Arabidopsis thaliana]
gi|22135950|gb|AAM91557.1| putative signal peptidase I
[Arabidopsis thaliana]
Length = 340
Score = 150 bits (379), Expect = 1e-35
Identities = 67/98 (68%), Positives = 82/98 (83%)
Frame = -1
Query: 622 YTDDDVFIKRIVAKEGDIVEVRDGHLIVNGVERNEKFILEPPSYEMKPTRVPENYVFVMG 443
Y+ +DVFIKRIVA EGD VEVRDG L VN + + E F+LEP SYEM+P VP+ YVFV+G
Sbjct: 229 YSSNDVFIKRIVASEGDWVEVRDGKLFVNDIVQEEDFVLEPMSYEMEPMFVPKGYVFVLG 288
Query: 442 DNRNNSYDSHVWGPLPAKNIIGRSVFRYWPPNRIADTI 329
DNRN S+DSH WGPLP +NI+GRSVFRYWPP++++DTI
Sbjct: 289 DNRNKSFDSHNWGPLPIENIVGRSVFRYWPPSKVSDTI 326
>ref|NP_180603.1| putative signal peptidase I; protein id: At2g30440.1 [Arabidopsis
thaliana] gi|25408074|pir||E84708 probable signal
peptidase I [imported] - Arabidopsis thaliana
gi|1946373|gb|AAB63091.1| putative signal peptidase I
[Arabidopsis thaliana]
Length = 250
Score = 150 bits (379), Expect = 1e-35
Identities = 67/98 (68%), Positives = 82/98 (83%)
Frame = -1
Query: 622 YTDDDVFIKRIVAKEGDIVEVRDGHLIVNGVERNEKFILEPPSYEMKPTRVPENYVFVMG 443
Y+ +DVFIKRIVA EGD VEVRDG L VN + + E F+LEP SYEM+P VP+ YVFV+G
Sbjct: 139 YSSNDVFIKRIVASEGDWVEVRDGKLFVNDIVQEEDFVLEPMSYEMEPMFVPKGYVFVLG 198
Query: 442 DNRNNSYDSHVWGPLPAKNIIGRSVFRYWPPNRIADTI 329
DNRN S+DSH WGPLP +NI+GRSVFRYWPP++++DTI
Sbjct: 199 DNRNKSFDSHNWGPLPIENIVGRSVFRYWPPSKVSDTI 236
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 538,174,524
Number of Sequences: 1393205
Number of extensions: 11814394
Number of successful extensions: 29159
Number of sequences better than 10.0: 273
Number of HSP's better than 10.0 without gapping: 28269
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 29059
length of database: 448,689,247
effective HSP length: 118
effective length of database: 284,291,057
effective search space used: 25017613016
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)