Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC008788A_C01 KMC008788A_c01
(777 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
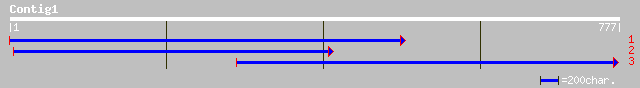
ref|NP_199366.1| putative protein; protein id: At5g45530.1 [Arab... 47 2e-04
ref|NP_199360.1| putative protein; protein id: At5g45470.1 [Arab... 44 0.003
ref|NP_193644.1| hypothetical protein; protein id: At4g19090.1 [... 42 0.013
ref|NP_199361.1| putative protein; protein id: At5g45480.1 [Arab... 41 0.022
ref|NP_199367.1| putative protein; protein id: At5g45540.1 [Arab... 40 0.049
>ref|NP_199366.1| putative protein; protein id: At5g45530.1 [Arabidopsis thaliana]
gi|9758743|dbj|BAB09181.1|
gene_id:MFC19.20~pir||T04430~similar to unknown protein
[Arabidopsis thaliana]
Length = 798
Score = 47.4 bits (111), Expect = 2e-04
Identities = 23/59 (38%), Positives = 35/59 (58%), Gaps = 1/59 (1%)
Frame = -3
Query: 427 KSILLWHIATYLCYNPVSCEE-ETYSERGLSFREGRKLLLEYMLYLIVMRTSMLPNEIG 254
+S+LLWHIAT LC+ + E S G RE K++ +YM+YL++MR ++ G
Sbjct: 595 QSLLLWHIATELCFQKEEGGKMEKLSREGYDDREFSKIISDYMMYLLIMRPKLMSEVAG 653
>ref|NP_199360.1| putative protein; protein id: At5g45470.1 [Arabidopsis thaliana]
gi|9758737|dbj|BAB09175.1|
gene_id:MFC19.14~pir||T04430~similar to unknown protein
[Arabidopsis thaliana]
Length = 866
Score = 43.9 bits (102), Expect = 0.003
Identities = 23/64 (35%), Positives = 38/64 (58%), Gaps = 6/64 (9%)
Frame = -3
Query: 427 KSILLWHIATYLCYNPVSCEEETYSE------RGLSFREGRKLLLEYMLYLIVMRTSMLP 266
+S+L+WHIAT LCY E+ET E + S RE K++ +YM+YL++++ ++
Sbjct: 662 QSLLMWHIATELCYQ--QHEKETIPEGYDEQRKHYSNREFSKIISDYMMYLLILQPGLMS 719
Query: 265 NEIG 254
G
Sbjct: 720 EVAG 723
>ref|NP_193644.1| hypothetical protein; protein id: At4g19090.1 [Arabidopsis
thaliana] gi|7487140|pir||T04430 hypothetical protein
T18B16.60 - Arabidopsis thaliana
gi|2832637|emb|CAA16766.1| hypothetical protein
[Arabidopsis thaliana] gi|7268704|emb|CAB78911.1|
hypothetical protein [Arabidopsis thaliana]
Length = 751
Score = 41.6 bits (96), Expect = 0.013
Identities = 20/58 (34%), Positives = 34/58 (58%), Gaps = 1/58 (1%)
Frame = -3
Query: 424 SILLWHIATYLCYNPV-SCEEETYSERGLSFREGRKLLLEYMLYLIVMRTSMLPNEIG 254
S+L+WHIAT LCY S +E + R+ K++ +YM+YL++M+ ++ G
Sbjct: 567 SLLIWHIATELCYQEEDSAKENCDKSEYHTNRKISKIISDYMMYLLIMQPKLMSEVAG 624
>ref|NP_199361.1| putative protein; protein id: At5g45480.1 [Arabidopsis thaliana]
gi|9758738|dbj|BAB09176.1|
gene_id:MFC19.15~pir||T04430~similar to unknown protein
[Arabidopsis thaliana]
Length = 877
Score = 40.8 bits (94), Expect = 0.022
Identities = 20/58 (34%), Positives = 35/58 (59%)
Frame = -3
Query: 427 KSILLWHIATYLCYNPVSCEEETYSERGLSFREGRKLLLEYMLYLIVMRTSMLPNEIG 254
+S+L+WHIAT L Y + +S R S K+L +YM+YL++M+ +++ +G
Sbjct: 673 QSLLVWHIATELLYQTKKGTKANHSAREFS-----KILSDYMMYLLMMQPTLMSAVVG 725
>ref|NP_199367.1| putative protein; protein id: At5g45540.1 [Arabidopsis thaliana]
gi|9758744|dbj|BAB09182.1|
gene_id:MFC19.21~pir||T04430~similar to unknown protein
[Arabidopsis thaliana]
Length = 803
Score = 39.7 bits (91), Expect = 0.049
Identities = 24/61 (39%), Positives = 39/61 (63%), Gaps = 8/61 (13%)
Frame = -3
Query: 427 KSILLWHIATYLCY-NPVS---CEEETYS----ERGLSFREGRKLLLEYMLYLIVMRTSM 272
+SILLWHIAT L Y P+ E+E +S + S RE K+L +YM+YL++++ ++
Sbjct: 594 QSILLWHIATELLYQKPIDKKVTEKEEHSTNREKEEHSNREFSKILSDYMMYLLIVQPTL 653
Query: 271 L 269
+
Sbjct: 654 M 654
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 619,867,811
Number of Sequences: 1393205
Number of extensions: 13019965
Number of successful extensions: 25536
Number of sequences better than 10.0: 19
Number of HSP's better than 10.0 without gapping: 24585
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 25517
length of database: 448,689,247
effective HSP length: 121
effective length of database: 280,111,442
effective search space used: 38375267554
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)