Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC008452A_C01 KMC008452A_c01
(651 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
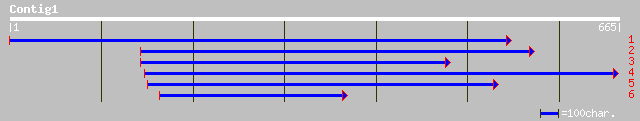
pir||E22845 hypothetical protein 4 - Trypanosoma brucei mitochon... 37 0.18
ref|NP_702802.1| hypothetical protein [Plasmodium falciparum 3D7... 37 0.30
gb|AAN86606.1| MURF1 [Trypanosoma sp.] 36 0.51
gb|EAA22516.1| hypothetical protein [Plasmodium yoelii yoelii] 35 1.1
ref|NP_050068.1| NH2 terminus uncertain [Leishmania tarentolae] ... 35 1.1
>pir||E22845 hypothetical protein 4 - Trypanosoma brucei mitochondrion
Length = 445
Score = 37.4 bits (85), Expect = 0.18
Identities = 28/103 (27%), Positives = 52/103 (50%), Gaps = 8/103 (7%)
Frame = -2
Query: 431 LIWICLLRRIRCLLPLVPSFWLS*-CCSISLASFC*CFMLYCVLFVVVVGTWFFSCFLSY 255
L+ CL I + P+V F++ C + L+ C C + +++++++ ++F ++
Sbjct: 341 LLLACLFLCIGAI-PIVFGFFIKVFCLLLQLSYLCICIGFFFIIWLIIIYIFYFRLIVNI 399
Query: 254 FSFFYPFLSLVDYKL*PIS*EG-----CS--HVLFISIIILFE 147
F F Y FL KL I+ + CS ++LF II LF+
Sbjct: 400 FIFSYQFLGFWVVKLSFINIKNLLFFICSSVYILFFDIINLFD 442
>ref|NP_702802.1| hypothetical protein [Plasmodium falciparum 3D7]
gi|23498218|emb|CAD49189.1| hypothetical protein
[Plasmodium falciparum 3D7]
Length = 104
Score = 36.6 bits (83), Expect = 0.30
Identities = 28/71 (39%), Positives = 37/71 (51%)
Frame = -1
Query: 375 FLAVLMLFNLVSLFLLMFYVVLCFVCSCRWNLVLFLFSFLFQFLLSFPKSCGL*TMTYLM 196
FL+ L+ F L L + +L F+ S L+ FL SFL FLLSF C L YL+
Sbjct: 33 FLSFLLSFLLSFLLSFLLSFLLSFLLSF---LLSFLLSFLLSFLLSFLLICFL---NYLL 86
Query: 195 RRVFSCSLYFN 163
FS SL+ +
Sbjct: 87 SFSFSFSLFLS 97
>gb|AAN86606.1| MURF1 [Trypanosoma sp.]
Length = 97
Score = 35.8 bits (81), Expect = 0.51
Identities = 22/82 (26%), Positives = 43/82 (51%), Gaps = 1/82 (1%)
Frame = -2
Query: 392 LPLVPSFWLS*CCSISLASFC*-CFMLYCVLFVVVVGTWFFSCFLSYFSFFYPFLSLVDY 216
+P+V F+L C + S+ C + +L+++VV ++F ++ F F Y F+
Sbjct: 5 IPIVFGFFLKVFCLLLQLSYLGVCVTFFLMLWLIVVYIFYFRLIVNIFIFSYQFIGFWVV 64
Query: 215 KL*PIS*EGCSHVLFISIIILF 150
+L ++ + VL I++ ILF
Sbjct: 65 RLYVVTFQNLFFVLAINLYILF 86
>gb|EAA22516.1| hypothetical protein [Plasmodium yoelii yoelii]
Length = 1668
Score = 34.7 bits (78), Expect = 1.1
Identities = 22/81 (27%), Positives = 39/81 (47%), Gaps = 1/81 (1%)
Frame = +2
Query: 194 LMR*VIVYSPQDLGKDKRN*NKKENKKRTKFQRQLQTKHNTT*NISKKR-LTRLNSIRTA 370
LM ++ ++G +R N + K+ +L +H+TT I KKR +N +
Sbjct: 620 LMNKNVIIDTVNVGNKRRGNNLQTKYSDEKYNNELSEEHDTTHLIDKKRKFNDINEHKNN 679
Query: 371 KKMGPVAASI*SCAVDISKLK 433
K G +I +C V+ +K+K
Sbjct: 680 SKNGKYNGTIKACDVNENKIK 700
>ref|NP_050068.1| NH2 terminus uncertain [Leishmania tarentolae] gi|83996|pir||B26696
hypothetical protein 1 (CYb-COII intergenic region) -
Leishmania tarentolae mitochondrion (fragment)
gi|896286|gb|AAA96601.1| NH2 terminus uncertain
Length = 443
Score = 34.7 bits (78), Expect = 1.1
Identities = 19/82 (23%), Positives = 41/82 (49%), Gaps = 1/82 (1%)
Frame = -2
Query: 392 LPLVPSFWLS*CCSISLASFC*-CFMLYCVLFVVVVGTWFFSCFLSYFSFFYPFLSLVDY 216
+P+V F+L C + S+ C + + +++++++ ++F ++ F F Y F+
Sbjct: 351 IPIVFGFFLKVFCLLLHLSYLGICIVFFSIIWLIIIYIFYFRLIINIFIFSYQFIGFWVV 410
Query: 215 KL*PIS*EGCSHVLFISIIILF 150
+L + +L SI ILF
Sbjct: 411 RLHILGFSNLFFILSFSIFILF 432
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 557,439,332
Number of Sequences: 1393205
Number of extensions: 11982493
Number of successful extensions: 35881
Number of sequences better than 10.0: 63
Number of HSP's better than 10.0 without gapping: 32705
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 35716
length of database: 448,689,247
effective HSP length: 118
effective length of database: 284,291,057
effective search space used: 27860523586
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)