Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
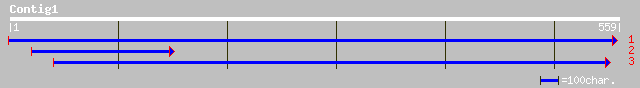
Query= KMC008402A_C01 KMC008402A_c01
(559 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAM91884.1| putative receptor-like protein kinase [Oryza sati... 129 3e-29
pir||E96602 hypothetical protein T6H22.9 [imported] - Arabidopsi... 79 4e-14
ref|NP_176008.1| wall-associated kinase 2, putative; protein id:... 79 4e-14
gb|AAF02839.1|AC009894_10 Similar to serine/threonine kinases [A... 75 4e-13
ref|NP_564710.1| receptor protein kinase, putative; protein id: ... 75 4e-13
>gb|AAM91884.1| putative receptor-like protein kinase [Oryza sativa (japonica
cultivar-group)]
Length = 373
Score = 129 bits (323), Expect = 3e-29
Identities = 70/132 (53%), Positives = 99/132 (74%)
Frame = -1
Query: 559 VVLEIISGQKSSELRDDADGEFLLQRAWKLYEVERHLELVDKTLNSEDYDGEEVKRIIEI 380
VVLEIISG+K ++ R D D ++LL+ AWKLYE +ELVDK+L+ ++Y+ EEVK+II+I
Sbjct: 245 VVLEIISGRKLNDARLDPDSQYLLEWAWKLYENNNLIELVDKSLDPKEYNPEEVKKIIQI 304
Query: 379 ALLCTQASASARPTMSEIVVLLKSKNLLVHMKPAMPVFVESNFRPRTDTSTSTGSSTSKS 200
ALLCTQ++ ++RPTMSE+VVLL +KN +P P F+++ R R +TS+S+ SS SK
Sbjct: 305 ALLCTQSAVASRPTMSEVVVLLLTKN-SSEFQPTRPTFIDAISRVRGETSSSSSSSASK- 362
Query: 199 NATVSTSVLSAR 164
AT+S + SAR
Sbjct: 363 -ATISITQYSAR 373
>pir||E96602 hypothetical protein T6H22.9 [imported] - Arabidopsis thaliana
gi|6056372|gb|AAF02836.1|AC009894_7 Very similar to
receptor-like serine/threonine kinase [Arabidopsis
thaliana]
Length = 858
Score = 79.0 bits (193), Expect = 4e-14
Identities = 50/131 (38%), Positives = 84/131 (63%), Gaps = 1/131 (0%)
Frame = -1
Query: 559 VVLEIISGQKSSELRDDADGEFLLQRAWKLYEVERHLELVDKTLNSEDYDGEEVKRIIEI 380
V LE++SG+K+S+ + ++LL+ AW L+E R +EL+D L+ +Y+ EEVKR+I I
Sbjct: 710 VALELVSGRKNSDENLEEGKKYLLEWAWNLHEKNRDVELIDDELS--EYNMEEVKRMIGI 767
Query: 379 ALLCTQASASARPTMSEIVVLLKSKNLLVHMKPAMPVFV-ESNFRPRTDTSTSTGSSTSK 203
ALLCTQ+S + RP MS +V +L S + V+ + P ++ + F DT++S+ S+
Sbjct: 768 ALLCTQSSYALRPPMSRVVAML-SGDAEVNDATSKPGYLTDCTF---DDTTSSSFSNFQT 823
Query: 202 SNATVSTSVLS 170
+ + STS ++
Sbjct: 824 KDTSFSTSFIA 834
>ref|NP_176008.1| wall-associated kinase 2, putative; protein id: At1g56120.1
[Arabidopsis thaliana]
Length = 1045
Score = 79.0 bits (193), Expect = 4e-14
Identities = 50/131 (38%), Positives = 84/131 (63%), Gaps = 1/131 (0%)
Frame = -1
Query: 559 VVLEIISGQKSSELRDDADGEFLLQRAWKLYEVERHLELVDKTLNSEDYDGEEVKRIIEI 380
V LE++SG+K+S+ + ++LL+ AW L+E R +EL+D L+ +Y+ EEVKR+I I
Sbjct: 897 VALELVSGRKNSDENLEEGKKYLLEWAWNLHEKNRDVELIDDELS--EYNMEEVKRMIGI 954
Query: 379 ALLCTQASASARPTMSEIVVLLKSKNLLVHMKPAMPVFV-ESNFRPRTDTSTSTGSSTSK 203
ALLCTQ+S + RP MS +V +L S + V+ + P ++ + F DT++S+ S+
Sbjct: 955 ALLCTQSSYALRPPMSRVVAML-SGDAEVNDATSKPGYLTDCTF---DDTTSSSFSNFQT 1010
Query: 202 SNATVSTSVLS 170
+ + STS ++
Sbjct: 1011 KDTSFSTSFIA 1021
>gb|AAF02839.1|AC009894_10 Similar to serine/threonine kinases [Arabidopsis thaliana]
Length = 889
Score = 75.5 bits (184), Expect = 4e-13
Identities = 38/82 (46%), Positives = 56/82 (67%)
Frame = -1
Query: 559 VVLEIISGQKSSELRDDADGEFLLQRAWKLYEVERHLELVDKTLNSEDYDGEEVKRIIEI 380
V LEI+SG+ +S D D ++LL+ AW L++ +R +E+VD L ++D EEVKR+I +
Sbjct: 753 VALEIVSGRPNSSPELDDDKQYLLEWAWSLHQEQRDMEVVDPDLT--EFDKEEVKRVIGV 810
Query: 379 ALLCTQASASARPTMSEIVVLL 314
A LCTQ + RPTMS +V +L
Sbjct: 811 AFLCTQTDHAIRPTMSRVVGML 832
>ref|NP_564710.1| receptor protein kinase, putative; protein id: At1g56145.1
[Arabidopsis thaliana]
Length = 1012
Score = 75.5 bits (184), Expect = 4e-13
Identities = 38/82 (46%), Positives = 56/82 (67%)
Frame = -1
Query: 559 VVLEIISGQKSSELRDDADGEFLLQRAWKLYEVERHLELVDKTLNSEDYDGEEVKRIIEI 380
V LEI+SG+ +S D D ++LL+ AW L++ +R +E+VD L ++D EEVKR+I +
Sbjct: 876 VALEIVSGRPNSSPELDDDKQYLLEWAWSLHQEQRDMEVVDPDLT--EFDKEEVKRVIGV 933
Query: 379 ALLCTQASASARPTMSEIVVLL 314
A LCTQ + RPTMS +V +L
Sbjct: 934 AFLCTQTDHAIRPTMSRVVGML 955
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 436,626,445
Number of Sequences: 1393205
Number of extensions: 8456446
Number of successful extensions: 25514
Number of sequences better than 10.0: 821
Number of HSP's better than 10.0 without gapping: 24040
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 25203
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 19808345223
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)