Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC008326A_C01 KMC008326A_c01
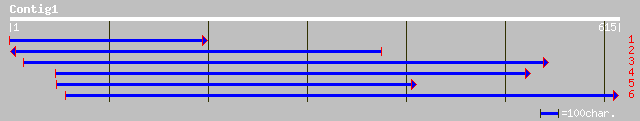
(610 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAG13524.1|AC068924_29 putative non-LTR retroelement reverse ... 68 1e-10
ref|NP_182048.1| putative non-LTR retroelement reverse transcrip... 58 8e-08
gb|AAO19364.1| hypothetical protein [Oryza sativa (japonica cult... 57 2e-07
ref|NP_680171.1| similar to reverse transcriptase, putative; pro... 49 4e-05
ref|NP_568371.1| reverse-transcriptase - like protein; protein i... 49 7e-05
>gb|AAG13524.1|AC068924_29 putative non-LTR retroelement reverse transcriptase [Oryza sativa
(japonica cultivar-group)]
Length = 1382
Score = 67.8 bits (164), Expect = 1e-10
Identities = 39/121 (32%), Positives = 64/121 (52%), Gaps = 2/121 (1%)
Frame = -2
Query: 549 WEPPPEGIIKVNIDAG-WSGSCSTGFSLVARSHNAVMLVAATHLEETQIEPTLAEALALR 373
W+PPP + +N DA +S S + G + R + LVA + + + P LAEALA+R
Sbjct: 1224 WQPPPASVWMINSDAAIFSSSRTMGVGALIRDNTGKCLVACSEMISDVVLPELAEALAIR 1283
Query: 372 WCLSMIKEMEMERIIVESYSLVVLNTLHQKTK-RPDIELVVYDSRELAAEFNFINFKHIG 196
L + KE +E I++ S L V+ + + R + V+ D ++LA+ F +F H+
Sbjct: 1284 RALGLAKEEGLEHIVMASDCLTVIRRIQTSGRDRSGVGCVIEDIKKLASTFVLCSFMHVN 1343
Query: 195 R 193
R
Sbjct: 1344 R 1344
>ref|NP_182048.1| putative non-LTR retroelement reverse transcriptase; protein id:
At2g45230.1 [Arabidopsis thaliana]
gi|25408936|pir||A84888 hypothetical protein At2g45230
[imported] - Arabidopsis thaliana
gi|2583130|gb|AAB82639.1| putative non-LTR retroelement
reverse transcriptase [Arabidopsis thaliana]
Length = 1374
Score = 58.2 bits (139), Expect = 8e-08
Identities = 35/127 (27%), Positives = 56/127 (43%), Gaps = 3/127 (2%)
Frame = -2
Query: 561 RRMVWEPPPEGIIKVNIDAGWS---GSCSTGFSLVARSHNAVMLVAATHLEETQIEPTLA 391
R + W+PP G +K N D WS G+C G+ V R+H +L +Q
Sbjct: 1211 RCVKWQPPSHGWVKCNTDGAWSKDLGNCGVGW--VLRNHTGRLLWLGLRALPSQQSVLET 1268
Query: 390 EALALRWCLSMIKEMEMERIIVESYSLVVLNTLHQKTKRPDIELVVYDSRELAAEFNFIN 211
E ALRW + + R+I ES S +++ + + P + + D R L F +
Sbjct: 1269 EVEALRWAVLSLSRFNYRRVIFESDSQYLVSLIQNEMDIPSLAPRIQDIRNLLRHFEEVK 1328
Query: 210 FKHIGRK 190
F+ R+
Sbjct: 1329 FQFTRRE 1335
>gb|AAO19364.1| hypothetical protein [Oryza sativa (japonica cultivar-group)]
Length = 499
Score = 57.0 bits (136), Expect = 2e-07
Identities = 32/84 (38%), Positives = 44/84 (52%), Gaps = 1/84 (1%)
Frame = -2
Query: 597 AQDRKESSIEAPRRMVWEPPPEGIIKVNIDAG-WSGSCSTGFSLVARSHNAVMLVAATHL 421
A R +SS PR W PPP G + +N DA + SC G + R H+ L+A
Sbjct: 411 AAPRGDSSPSIPR---WTPPPVGTVMINCDAALFQSSCQMGIGFLIRDHDGRCLLALNER 467
Query: 420 EETQIEPTLAEALALRWCLSMIKE 349
+ +P LAEA+A+R LS+ KE
Sbjct: 468 VQNVTQPELAEAVAIRRALSLAKE 491
>ref|NP_680171.1| similar to reverse transcriptase, putative; protein id: At5g17725.1
[Arabidopsis thaliana]
Length = 721
Score = 49.3 bits (116), Expect = 4e-05
Identities = 30/125 (24%), Positives = 54/125 (43%), Gaps = 4/125 (3%)
Frame = -2
Query: 609 EWTDAQD-RKESSIEAPRRMVWEPPPEGIIKVNIDAGW---SGSCSTGFSLVARSHNAVM 442
EW D +S ++P + W+PP G K N+DA W S C G+ + R
Sbjct: 536 EWVDKPVISPKSPQQSPTPVTWQPPSPGTYKCNVDAAWKAESDRCGVGW--ILRDATGAA 593
Query: 441 LVAATHLEETQIEPTLAEALALRWCLSMIKEMEMERIIVESYSLVVLNTLHQKTKRPDIE 262
+ + + AEA AL W + + + + ++ E+ S V++ L++ P +
Sbjct: 594 KWVGSKAYPSLVSSLEAEATALTWAMRCLDNLGVTEVVFETDSQVLVKALNEPEMWPRLS 653
Query: 261 LVVYD 247
+ D
Sbjct: 654 SYIDD 658
>ref|NP_568371.1| reverse-transcriptase - like protein; protein id: At5g19270.1
[Arabidopsis thaliana]
Length = 365
Score = 48.5 bits (114), Expect = 7e-05
Identities = 35/144 (24%), Positives = 61/144 (42%), Gaps = 8/144 (5%)
Frame = -2
Query: 549 WEPPPEGIIKVNIDAGW-SGSCSTGFSLVARSHNAVMLVAATHLEETQIEPTLAEALALR 373
W PP GI+K N++A W + +G + +AR N + L A +AE +
Sbjct: 61 WTPPDFGIVKCNVNAHWGNADMMSGVAWIARDVNGLALYHARDAFTRSSNRIMAELRCIL 120
Query: 372 WCLSMIKEMEMERIIVESYSLVVLNTLHQKTKRPDIELVVYDSRELAAEFNFINFK---- 205
W + ++++ ++ +I+ S + L+ P + + F FI F+
Sbjct: 121 WVVKSLRDLRLQNVIISSDCQAAIAALYNPRDWPRYAMWLDTIFIACQSFPFIIFELVSC 180
Query: 204 ---HIGRKVINSGTCFGRFSC*LS 142
I R + S T GRF+ LS
Sbjct: 181 PANSIARDIAESVTRDGRFNPYLS 204
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 502,747,034
Number of Sequences: 1393205
Number of extensions: 10199613
Number of successful extensions: 25633
Number of sequences better than 10.0: 58
Number of HSP's better than 10.0 without gapping: 24965
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 25621
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 24283162270
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)