Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC008277A_C01 KMC008277A_c01
(578 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
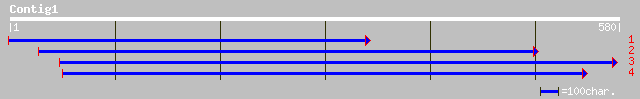
ref|NP_199849.1| putative protein; protein id: At5g50380.1 [Arab... 35 0.52
ref|NP_189039.1| hypothetical protein; protein id: At3g23970.1 [... 34 1.2
gb|EAA41029.1| GLP_12_49813_66465 [Giardia lamblia ATCC 50803] 33 2.6
gb|EAA21325.1| hypothetical protein [Plasmodium yoelii yoelii] 33 2.6
>ref|NP_199849.1| putative protein; protein id: At5g50380.1 [Arabidopsis thaliana]
gi|9758920|dbj|BAB09457.1|
gb|AAD25781.1~gene_id:MXI22.10~strong similarity to
unknown protein [Arabidopsis thaliana]
Length = 637
Score = 35.4 bits (80), Expect = 0.52
Identities = 17/49 (34%), Positives = 25/49 (50%), Gaps = 2/49 (4%)
Frame = -2
Query: 304 VYSKSQRSILDGCLWLCTKSSFSLSRLRR--WTRLREKMHSWMAIAKIT 164
VYS +R LD CL + S+ +++ W + EKM W+ KIT
Sbjct: 219 VYSSVRRDALDDCLMILGVEKLSIEEVQKIDWKSMDEKMKKWIQAVKIT 267
>ref|NP_189039.1| hypothetical protein; protein id: At3g23970.1 [Arabidopsis
thaliana] gi|9294666|dbj|BAB03015.1|
gene_id:F14O13.16~pir||T05437~similar to unknown protein
[Arabidopsis thaliana]
Length = 413
Score = 34.3 bits (77), Expect = 1.2
Identities = 20/72 (27%), Positives = 36/72 (49%), Gaps = 1/72 (1%)
Frame = -3
Query: 531 FHDHNSIHLFVWKMVKY-GALESWTQLLSISYERLHCIGFPYIPVPMFLLEDGDILMIAI 355
F ++ + L VW++VKY G E+W +S + +G Y PV M L+ +I+ +
Sbjct: 280 FGENGNRRLRVWRLVKYTGGPEAWQLFWEVSLASVTKLGIDYFPVVMHPLK-SEIIYLWS 338
Query: 354 TENLEFIKYNIR 319
+ +N+R
Sbjct: 339 RNKKGMVLFNLR 350
>gb|EAA41029.1| GLP_12_49813_66465 [Giardia lamblia ATCC 50803]
Length = 5550
Score = 33.1 bits (74), Expect = 2.6
Identities = 21/82 (25%), Positives = 35/82 (42%), Gaps = 1/82 (1%)
Frame = -3
Query: 537 YLFHDHNSIHLFVWKMVKYGALESWTQLLSISYERLHCIGFPYIPVPMFLLEDGDILMIA 358
+LF HN + K V G + F Y P P+ L+E+ ++
Sbjct: 5126 FLFDTHNPFSFYYKKAVAQGDTD-----------------FDYAPPPIVLIENSGNQVVV 5168
Query: 357 ITENL-EFIKYNIRDYSMEYIP 295
+TE + + +KY+IR Y +P
Sbjct: 5169 LTEMITKLVKYSIRKYGAAGLP 5190
>gb|EAA21325.1| hypothetical protein [Plasmodium yoelii yoelii]
Length = 2112
Score = 33.1 bits (74), Expect = 2.6
Identities = 21/74 (28%), Positives = 33/74 (44%), Gaps = 8/74 (10%)
Frame = -1
Query: 200 ENALLDGNCQDHINSQTIFDLYLISIMKSLK--------FGEYFKLCCHGSEYYICLGKD 45
EN+L D N + INS + ++YL + K F ++ + S+YY C
Sbjct: 814 ENSLRDNNNYNSINSNIVTNMYLSNNSKKYNSVSNSCHLFNNKNEMSNNSSKYYACKNGP 873
Query: 44 IVILRNSNILIPSF 3
+L SN L P +
Sbjct: 874 YNVLNKSNTLHPYY 887
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 489,213,080
Number of Sequences: 1393205
Number of extensions: 10374938
Number of successful extensions: 25418
Number of sequences better than 10.0: 8
Number of HSP's better than 10.0 without gapping: 24652
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 25412
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 21426319650
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)