Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
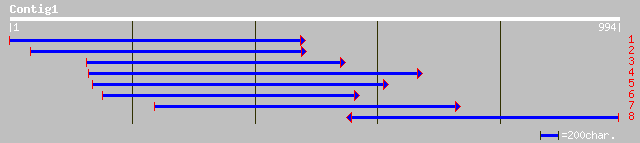
Query= KMC008270A_C01 KMC008270A_c01
(984 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
emb|CAD29731.1| protease inhibitor [Sesbania rostrata] 339 3e-92
prf||1802409A albumin 96 1e-18
sp|P32765|ASP_THECC 21 kDa seed protein precursor gi|99954|pir||... 96 1e-18
gb|AAL85644.1| trypsin inhibitor [Theobroma subincanum] 86 9e-16
gb|AAL85648.1| trypsin inhibitor [Theobroma obovatum] gi|1917170... 86 1e-15
>emb|CAD29731.1| protease inhibitor [Sesbania rostrata]
Length = 215
Score = 339 bits (870), Expect = 3e-92
Identities = 163/219 (74%), Positives = 188/219 (85%)
Frame = -2
Query: 965 LKVAASMFNPSMFFLSLLILNTQPLLAAEPEPEPVVDKQGNPLEPGVGYYAWPVWADKGG 786
+KVA F P + FLSLL+ NT PLLAAEPEP VVDKQG PL+PGVGYY WP+WAD GG
Sbjct: 1 MKVAKLQFLP-LCFLSLLVFNTHPLLAAEPEP--VVDKQGEPLQPGVGYYVWPLWADMGG 57
Query: 785 LTLGRTRNKTCPLDIIRDPSKIGTPIELHVADSESGYIPTLTDLTVEMPILGSRCQEPKV 606
LTLG+TRNKTCPLD+IRDPS IG+P+ HVA S+ G+IPTLTDLT+ +PILGS C+EPKV
Sbjct: 58 LTLGQTRNKTCPLDVIRDPSFIGSPVTFHVAGSDLGFIPTLTDLTINIPILGSHCKEPKV 117
Query: 605 WRLLKVGSGFWFLSTGGVAGNLVSKFKIERLAGEHAYEIYSFKFCPSVPGVLCAPVGTYE 426
WRL K GSGFWF+STGGVAG+L+SKFKIERL GEHAYEIYSFKFCPSVPGVLCAPVGT+E
Sbjct: 118 WRLSKEGSGFWFVSTGGVAGDLISKFKIERLEGEHAYEIYSFKFCPSVPGVLCAPVGTFE 177
Query: 425 DADGTQVMAIGDDLETYYVRFQKASSTLAQKKNPDLSRV 309
DADGT+VMA+GD +E YYVRFQK S++ +QKK + S V
Sbjct: 178 DADGTKVMAVGDGIEPYYVRFQK-STSYSQKKGQEFSSV 215
>prf||1802409A albumin
Length = 221
Score = 95.5 bits (236), Expect = 1e-18
Identities = 71/195 (36%), Positives = 102/195 (51%), Gaps = 15/195 (7%)
Frame = -2
Query: 866 PVVDKQGNPLEPGVGYYAWP--VWADKGGLTLGRTRNKTCPLDIIRDPSKI--GTPIELH 699
PV+D G+ L+ GV YY A GGL LGR ++CP +++ S + GTP+
Sbjct: 30 PVLDTDGDELQTGVQYYVLSSISGAGGGGLALGRATGQSCPEIVVQRRSDLDNGTPVIFS 89
Query: 698 VADSESGYIPTLTDLTVE-MPILGSRCQEPKVWRL--LKVGSGFWFLSTGGVAG-----N 543
ADS+ + TD+ +E +PI C VWRL +G W+++T GV G
Sbjct: 90 NADSKDDVVRVSTDVNIEFVPIRDRLCSTSTVWRLDNYDNSAGKWWVTTDGVKGEPGPNT 149
Query: 542 LVSKFKIERLAGEHAYEIYSFKFCPSVPG---VLCAPVGTYEDADGTQVMAIGDDLETYY 372
L S FKIE+ AG Y+ F+FCPSV LC+ +G + D DG +A+ D+ +
Sbjct: 150 LCSWFKIEK-AGVLGYK---FRFCPSVCDSCTTLCSDIGRHSDDDGQIRLALSDN--EWA 203
Query: 371 VRFQKASSTLAQKKN 327
F+KAS T+ Q N
Sbjct: 204 WMFKKASKTIKQVVN 218
>sp|P32765|ASP_THECC 21 kDa seed protein precursor gi|99954|pir||S16252 trypsin
inhibitor homolog - soybean gi|21909|emb|CAA39860.1| 21
kDa seed protein [Theobroma cacao]
Length = 221
Score = 95.5 bits (236), Expect = 1e-18
Identities = 71/195 (36%), Positives = 102/195 (51%), Gaps = 15/195 (7%)
Frame = -2
Query: 866 PVVDKQGNPLEPGVGYYAWP--VWADKGGLTLGRTRNKTCPLDIIRDPSKI--GTPIELH 699
PV+D G+ L+ GV YY A GGL LGR ++CP +++ S + GTP+
Sbjct: 30 PVLDTDGDELQTGVQYYVLSSISGAGGGGLALGRATGQSCPEIVVQRRSDLDNGTPVIFS 89
Query: 698 VADSESGYIPTLTDLTVE-MPILGSRCQEPKVWRL--LKVGSGFWFLSTGGVAG-----N 543
ADS+ + TD+ +E +PI C VWRL +G W+++T GV G
Sbjct: 90 NADSKDDVVRVSTDVNIEFVPIRDRLCSTSTVWRLDNYDNSAGKWWVTTDGVKGEPGPNT 149
Query: 542 LVSKFKIERLAGEHAYEIYSFKFCPSVPG---VLCAPVGTYEDADGTQVMAIGDDLETYY 372
L S FKIE+ AG Y+ F+FCPSV LC+ +G + D DG +A+ D+ +
Sbjct: 150 LCSWFKIEK-AGVLGYK---FRFCPSVCDSCTTLCSDIGRHSDDDGQIRLALSDN--EWA 203
Query: 371 VRFQKASSTLAQKKN 327
F+KAS T+ Q N
Sbjct: 204 WMFKKASKTIKQVVN 218
>gb|AAL85644.1| trypsin inhibitor [Theobroma subincanum]
Length = 154
Score = 85.9 bits (211), Expect = 9e-16
Identities = 58/158 (36%), Positives = 82/158 (51%), Gaps = 15/158 (9%)
Frame = -2
Query: 863 VVDKQGNPLEPGVGYYAWP-VW-ADKGGLTLGRTRNKTCPLDIIRDPSKI--GTPIELHV 696
V+D G+ L GV YY +W A GGL LGR ++ CP +++ S + GTP+
Sbjct: 1 VLDTDGDELRTGVQYYVMSAIWGAGGGGLALGRATDQKCPEIVVQRRSDLDNGTPVIFSN 60
Query: 695 ADSESGYIPTLTDLTVE-MPILGSRCQEPKVWRL--LKVGSGFWFLSTGGVAG-----NL 540
ADSE + TD+ +E +PI C VW+L +G W+++TGGV G L
Sbjct: 61 ADSEDDVVRVSTDINIEFVPIRDRLCSTSTVWKLDDYDNSAGKWWVTTGGVKGEPGPNTL 120
Query: 539 VSKFKIERLAGEHAYEIYSFKFCPSV---PGVLCAPVG 435
+ FKIE G +Y F+FCPSV LC+ +G
Sbjct: 121 SNWFKIEEAGG----NVYKFRFCPSVCDSCATLCSDIG 154
>gb|AAL85648.1| trypsin inhibitor [Theobroma obovatum] gi|19171709|gb|AAL85649.1|
trypsin inhibitor [Theobroma obovatum]
Length = 154
Score = 85.5 bits (210), Expect = 1e-15
Identities = 59/158 (37%), Positives = 82/158 (51%), Gaps = 15/158 (9%)
Frame = -2
Query: 863 VVDKQGNPLEPGVGYYAWPV-W-ADKGGLTLGRTRNKTCPLDIIRDPSKI--GTPIELHV 696
V+D G+ L GV YY V W A GGL LGR ++ CP +++ S + GTP+
Sbjct: 1 VLDTDGDELRTGVQYYVVSVIWGAGGGGLALGRATDQKCPEIVVQRRSDLDNGTPVIFSN 60
Query: 695 ADSESGYIPTLTDLTVE-MPILGSRCQEPKVWRL--LKVGSGFWFLSTGGVAG-----NL 540
ADSE + TD+ +E +PI C VW+L +G W+++TGGV G L
Sbjct: 61 ADSEDDVVRVSTDINIEFVPIRDRLCSTSTVWKLDDYDNSAGKWWVTTGGVKGEPGPNTL 120
Query: 539 VSKFKIERLAGEHAYEIYSFKFCPSV---PGVLCAPVG 435
+ FKIE G +Y F+FCPSV LC+ +G
Sbjct: 121 SNWFKIEEAGG----NVYKFRFCPSVCDSCATLCSDIG 154
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 903,549,683
Number of Sequences: 1393205
Number of extensions: 21670939
Number of successful extensions: 57068
Number of sequences better than 10.0: 165
Number of HSP's better than 10.0 without gapping: 51948
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 56666
length of database: 448,689,247
effective HSP length: 123
effective length of database: 277,325,032
effective search space used: 56574306528
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)