Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC008211A_C01 KMC008211A_c01
(853 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
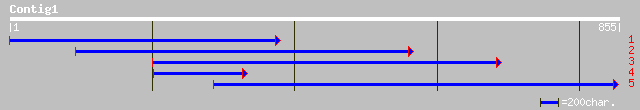
ref|NP_196457.2| putative protein; protein id: At5g08400.1, supp... 273 2e-72
emb|CAC08340.1| putative protein [Arabidopsis thaliana] 273 2e-72
ref|NP_194669.1| putative protein; protein id: At4g29400.1 [Arab... 153 3e-36
gb|ZP_00108378.1| hypothetical protein [Nostoc punctiforme] 114 1e-24
dbj|BAB85388.1| B1064G04.9 [Oryza sativa (japonica cultivar-grou... 113 3e-24
>ref|NP_196457.2| putative protein; protein id: At5g08400.1, supported by cDNA:
gi_18086462 [Arabidopsis thaliana]
gi|18086463|gb|AAL57685.1| unknown protein [Arabidopsis
thaliana] gi|24797008|gb|AAN64516.1| At5g08400/F8L15_130
[Arabidopsis thaliana]
Length = 333
Score = 273 bits (699), Expect = 2e-72
Identities = 123/148 (83%), Positives = 141/148 (95%)
Frame = -1
Query: 853 SFRDFNPTDSYIWFELYGSPSDQDVDLIGSVIQSWYVMGRLGSFNSSNLQMSNSSMEHDP 674
+FRDFNP DS+IWFELYG+PSD+DVDLIGSVIQ+WYVMGRLG+FN+SNLQ++N+S+E+DP
Sbjct: 186 NFRDFNPVDSFIWFELYGTPSDRDVDLIGSVIQAWYVMGRLGAFNTSNLQLANTSLEYDP 245
Query: 673 LYDAEQGFKVMPSSFHDISDIEFQDNWGRVWVDLGTCDYLAIDVLLNCLTVLSSEYLGIQ 494
LYDAE+GFKVMPSSFHDISD+EFQDNWGRVWVDLGT D A+DVLLNCLTV+SSEYLGIQ
Sbjct: 246 LYDAEKGFKVMPSSFHDISDVEFQDNWGRVWVDLGTSDIFALDVLLNCLTVMSSEYLGIQ 305
Query: 493 QVVFGGRCMGDWEEGMTDPGHGYKYFKI 410
QVVFGG+ MGDWEEGMT+P GYKYFKI
Sbjct: 306 QVVFGGKRMGDWEEGMTNPDFGYKYFKI 333
>emb|CAC08340.1| putative protein [Arabidopsis thaliana]
Length = 287
Score = 273 bits (699), Expect = 2e-72
Identities = 123/148 (83%), Positives = 141/148 (95%)
Frame = -1
Query: 853 SFRDFNPTDSYIWFELYGSPSDQDVDLIGSVIQSWYVMGRLGSFNSSNLQMSNSSMEHDP 674
+FRDFNP DS+IWFELYG+PSD+DVDLIGSVIQ+WYVMGRLG+FN+SNLQ++N+S+E+DP
Sbjct: 140 NFRDFNPVDSFIWFELYGTPSDRDVDLIGSVIQAWYVMGRLGAFNTSNLQLANTSLEYDP 199
Query: 673 LYDAEQGFKVMPSSFHDISDIEFQDNWGRVWVDLGTCDYLAIDVLLNCLTVLSSEYLGIQ 494
LYDAE+GFKVMPSSFHDISD+EFQDNWGRVWVDLGT D A+DVLLNCLTV+SSEYLGIQ
Sbjct: 200 LYDAEKGFKVMPSSFHDISDVEFQDNWGRVWVDLGTSDIFALDVLLNCLTVMSSEYLGIQ 259
Query: 493 QVVFGGRCMGDWEEGMTDPGHGYKYFKI 410
QVVFGG+ MGDWEEGMT+P GYKYFKI
Sbjct: 260 QVVFGGKRMGDWEEGMTNPDFGYKYFKI 287
>ref|NP_194669.1| putative protein; protein id: At4g29400.1 [Arabidopsis thaliana]
gi|25407612|pir||A85343 hypothetical protein AT4g29400
[imported] - Arabidopsis thaliana
gi|7269838|emb|CAB79698.1| putative protein [Arabidopsis
thaliana]
Length = 288
Score = 153 bits (387), Expect = 3e-36
Identities = 67/148 (45%), Positives = 103/148 (69%)
Frame = -1
Query: 853 SFRDFNPTDSYIWFELYGSPSDQDVDLIGSVIQSWYVMGRLGSFNSSNLQMSNSSMEHDP 674
+F F ++SY+W E Y +P D+D+ LI I+SW+++GRLG +NS N+Q+S + ++ P
Sbjct: 141 TFNKFEYSNSYMWLEFYNTPLDKDIALISDTIRSWHILGRLGGYNSMNMQLSQAPLDKRP 200
Query: 673 LYDAEQGFKVMPSSFHDISDIEFQDNWGRVWVDLGTCDYLAIDVLLNCLTVLSSEYLGIQ 494
YDA G V P++F++I D+E QDN R+W+D+GT + L +DVL+N LT +SS+Y+GI+
Sbjct: 201 NYDAILGANVEPTTFYNIGDLEVQDNVARIWLDIGTSEPLILDVLINALTQISSDYVGIK 260
Query: 493 QVVFGGRCMGDWEEGMTDPGHGYKYFKI 410
+VVFGG W+E MT G++ KI
Sbjct: 261 KVVFGGSEFESWKENMTSEESGFRVHKI 288
>gb|ZP_00108378.1| hypothetical protein [Nostoc punctiforme]
Length = 150
Score = 114 bits (286), Expect = 1e-24
Identities = 52/132 (39%), Positives = 84/132 (63%), Gaps = 1/132 (0%)
Frame = -1
Query: 850 FRDFNPTDSYIWFELYGSPSDQDVDLIGSVIQSWYVMGRLGSFNSSNLQMSNSSMEHDPL 671
FR+ NP D +IW + +PS ++ + V SW+ +G+LG+FN+ NLQ+ ++ ++ +
Sbjct: 5 FREINPFDMWIWLKFSTNPSAREKQYVEEVFNSWFYLGKLGAFNAENLQVQDTGLDISYM 64
Query: 670 YDAEQGF-KVMPSSFHDISDIEFQDNWGRVWVDLGTCDYLAIDVLLNCLTVLSSEYLGIQ 494
EQG+ K + + H+I + E++ WGR W DLGT D +A+D+L+N LT LS EY+ I+
Sbjct: 65 NYDEQGYDKSLLALMHNIGEFEYEGQWGRCWFDLGTSDAIALDILINALTQLSQEYVTIE 124
Query: 493 QVVFGGRCMGDW 458
Q+ GG DW
Sbjct: 125 QLYIGGE-NPDW 135
>dbj|BAB85388.1| B1064G04.9 [Oryza sativa (japonica cultivar-group)]
gi|21328094|dbj|BAC00678.1| B1144D11.25 [Oryza sativa
(japonica cultivar-group)]
Length = 338
Score = 113 bits (283), Expect = 3e-24
Identities = 53/117 (45%), Positives = 80/117 (68%), Gaps = 1/117 (0%)
Frame = -1
Query: 850 FRDFNPTDSYIWFELYGSPSDQDVDLIGSVIQSWYVMGRLGSFNSSNLQMSNSSME-HDP 674
F F+ ++SYIWFE Y + +DV LI ++SW+++GRLG NS N+Q+S ++ P
Sbjct: 134 FNKFDFSNSYIWFEFYHALLPKDVTLICDALRSWHIVGRLGGCNSMNMQLSQLPLDCQRP 193
Query: 673 LYDAEQGFKVMPSSFHDISDIEFQDNWGRVWVDLGTCDYLAIDVLLNCLTVLSSEYL 503
YDA +G P+SF++I D+E QDN RVWVD+G + L +D+LLN LT ++S++L
Sbjct: 194 TYDALEGANTTPTSFYNIGDLEIQDNIARVWVDIGIHEPLLLDILLNALTTINSDFL 250
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 752,060,539
Number of Sequences: 1393205
Number of extensions: 16973902
Number of successful extensions: 44526
Number of sequences better than 10.0: 62
Number of HSP's better than 10.0 without gapping: 39756
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 43250
length of database: 448,689,247
effective HSP length: 122
effective length of database: 278,718,237
effective search space used: 44873636157
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)