Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC008197A_C01 KMC008197A_c01
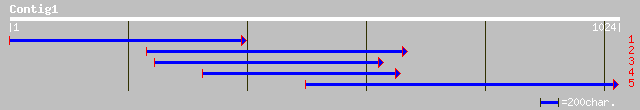
(1020 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAG13629.1|AC078840_20 putative steroid membrane binding prot... 218 2e-55
ref|NP_190458.1| putative progesterone-binding protein homolog A... 210 2e-53
gb|AAD34616.1|AF153284_1 putative progesterone-binding protein h... 208 9e-53
ref|NP_200037.1| progesterone-binding protein-like; protein id: ... 208 1e-52
gb|AAD34615.1|AF153283_1 putative progesterone-binding protein h... 204 1e-51
>gb|AAG13629.1|AC078840_20 putative steroid membrane binding protein [Oryza sativa (japonica
cultivar-group)]
Length = 232
Score = 218 bits (554), Expect = 2e-55
Identities = 116/175 (66%), Positives = 130/175 (74%), Gaps = 1/175 (0%)
Frame = -3
Query: 1015 EPLRPPVQIGEVTEEELKSYDGSDPEKPLLMAIKGQIYDVSQSRMFYGPGGPYALFAGKD 836
EPL PPVQ+GEV+EEEL+ YDGSDP+KPLLMAIKGQIYDV+QSRMFYGPGGPYALFAGKD
Sbjct: 63 EPLPPPVQLGEVSEEELRQYDGSDPKKPLLMAIKGQIYDVTQSRMFYGPGGPYALFAGKD 122
Query: 835 ASRALAKMSFEEKDLTGDISGLGSFELDALQDWEYKFMSKYVKVGTIKTTVTVTAPESTT 656
ASRALAKMSFE +DLTGDISGLG FELDALQDWEYKFM KYVKVGT+K TV V
Sbjct: 123 ASRALAKMSFEPQDLTGDISGLGPFELDALQDWEYKFMGKYVKVGTVKKTVPV-----ED 177
Query: 655 GEPSES-TTTPDADADDDDASKPTENGPSETEAVKSDETPSNIDAAKE*KGLDNS 494
G PS S TT A A + + + TE P E + + E + AA +G S
Sbjct: 178 GAPSTSPETTETAAAAEPEKAPATEEKPREVSSEEVKEKEDAVAAAAPDEGAKES 232
>ref|NP_190458.1| putative progesterone-binding protein homolog Atmp2; protein id:
At3g48890.1, supported by cDNA: gi_16930426, supported by
cDNA: gi_19310508, supported by cDNA: gi_4960153
[Arabidopsis thaliana] gi|11358660|pir||T49285 probable
progesterone-binding protein homolog Atmp2 - Arabidopsis
thaliana gi|7576227|emb|CAB87917.1| putative
progesterone-binding protein homolog Atmp2 [Arabidopsis
thaliana] gi|16930427|gb|AAL31899.1|AF419567_1
AT3g48890/T21J18_160 [Arabidopsis thaliana]
gi|19310509|gb|AAL84988.1| AT3g48890/T21J18_160
[Arabidopsis thaliana]
Length = 233
Score = 210 bits (535), Expect = 2e-53
Identities = 114/175 (65%), Positives = 132/175 (75%), Gaps = 8/175 (4%)
Frame = -3
Query: 1015 EPLRPPVQIGEVTEEELKSYDGSDPEKPLLMAIKGQIYDVSQSRMFYGPGGPYALFAGKD 836
EPL PPVQ+GE+TEEELK YDGSD +KPLLMAIKGQIYDVSQSRMFYGPGGPYALFAGKD
Sbjct: 60 EPLPPPVQLGEITEEELKLYDGSDSKKPLLMAIKGQIYDVSQSRMFYGPGGPYALFAGKD 119
Query: 835 ASRALAKMSFEEKDLTGDISGLGSFELDALQDWEYKFMSKYVKVGTI-------KTTVTV 677
ASRALAKMSFE++DLTGDISGLG+FEL+ALQDWEYKFMSKYVKVGTI K +
Sbjct: 120 ASRALAKMSFEDQDLTGDISGLGAFELEALQDWEYKFMSKYVKVGTIQKKDGEGKESSEP 179
Query: 676 TAPESTTGEPSESTTTPDADA-DDDDASKPTENGPSETEAVKSDETPSNIDAAKE 515
+ ++ + E + T +A A D+ S+ T +ET K D + DAAKE
Sbjct: 180 SEAKTASAEGLSTNTGEEASAITHDETSRSTGEKIAET-TEKKDVATDDDDAAKE 233
>gb|AAD34616.1|AF153284_1 putative progesterone-binding protein homolog [Arabidopsis thaliana]
gi|21555444|gb|AAM63860.1| progesterone-binding
protein-like [Arabidopsis thaliana]
Length = 220
Score = 208 bits (530), Expect = 9e-53
Identities = 104/149 (69%), Positives = 115/149 (76%), Gaps = 6/149 (4%)
Frame = -3
Query: 1012 PLRPPVQIGEVTEEELKSYDGSDPEKPLLMAIKGQIYDVSQSRMFYGPGGPYALFAGKDA 833
P+ PVQ+GE+TEEELK YDGSDP+KPLLMAIK QIYDV+QSRMFYGPGGPYALFAGKDA
Sbjct: 65 PIPQPVQVGEITEEELKQYDGSDPQKPLLMAIKHQIYDVTQSRMFYGPGGPYALFAGKDA 124
Query: 832 SRALAKMSFEEKDLTGDISGLGSFELDALQDWEYKFMSKYVKVGTIKT------TVTVTA 671
SRALAKMSFEEKDLT DISGLG FELDALQDWEYKFMSKY KVGT+K T +V+
Sbjct: 125 SRALAKMSFEEKDLTWDISGLGPFELDALQDWEYKFMSKYAKVGTVKVAGSEPETASVSE 184
Query: 670 PESTTGEPSESTTTPDADADDDDASKPTE 584
P + + TTTP+ D P E
Sbjct: 185 PTENVEQDAHVTTTPEKTVVDKSDDAPAE 213
>ref|NP_200037.1| progesterone-binding protein-like; protein id: At5g52240.1, supported
by cDNA: 28003., supported by cDNA: gi_15028112,
supported by cDNA: gi_4960155 [Arabidopsis thaliana]
gi|8885537|dbj|BAA97467.1| progesterone-binding
protein-like [Arabidopsis thaliana]
gi|24030304|gb|AAN41322.1| putative progesterone-binding
protein [Arabidopsis thaliana]
Length = 220
Score = 208 bits (529), Expect = 1e-52
Identities = 109/166 (65%), Positives = 120/166 (71%)
Frame = -3
Query: 1012 PLRPPVQIGEVTEEELKSYDGSDPEKPLLMAIKGQIYDVSQSRMFYGPGGPYALFAGKDA 833
P+ PVQ+GE+TEEELK YDGSDP+KPLLMAIK QIYDV+QSRMFYGPGGPYALFAGKDA
Sbjct: 65 PIPQPVQVGEITEEELKQYDGSDPQKPLLMAIKHQIYDVTQSRMFYGPGGPYALFAGKDA 124
Query: 832 SRALAKMSFEEKDLTGDISGLGSFELDALQDWEYKFMSKYVKVGTIKTTVTVTAPESTTG 653
SRALAKMSFEEKDLT D+SGLG FELDALQDWEYKFMSKY KVGT+K V E T
Sbjct: 125 SRALAKMSFEEKDLTWDVSGLGPFELDALQDWEYKFMSKYAKVGTVK----VAGSEPETA 180
Query: 652 EPSESTTTPDADADDDDASKPTENGPSETEAVKSDETPSNIDAAKE 515
SE T + DA P +T KSD+ P+ KE
Sbjct: 181 SVSEPTENVEQDAH-------VTTTPGKTVVDKSDDAPAETVLKKE 219
>gb|AAD34615.1|AF153283_1 putative progesterone-binding protein homolog [Arabidopsis thaliana]
Length = 253
Score = 204 bits (520), Expect = 1e-51
Identities = 102/128 (79%), Positives = 112/128 (86%)
Frame = -3
Query: 1015 EPLRPPVQIGEVTEEELKSYDGSDPEKPLLMAIKGQIYDVSQSRMFYGPGGPYALFAGKD 836
EPL PPVQ+GE+TEEELK YDGSD +KPLLMAIKGQIYDVSQSRMFYGPGGPYALFAGKD
Sbjct: 60 EPLPPPVQLGEITEEELKLYDGSDSKKPLLMAIKGQIYDVSQSRMFYGPGGPYALFAGKD 119
Query: 835 ASRALAKMSFEEKDLTGDISGLGSFELDALQDWEYKFMSKYVKVGTIKTTVTVTAPESTT 656
ASRALAKMSFE++DLTGDISGLG+FEL+ALQDWEYKFMSKYVKVGTI+ +
Sbjct: 120 ASRALAKMSFEDQDLTGDISGLGAFELEALQDWEYKFMSKYVKVGTIQKK---DGEGKES 176
Query: 655 GEPSESTT 632
EPSE+ T
Sbjct: 177 SEPSEAKT 184
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 879,855,608
Number of Sequences: 1393205
Number of extensions: 19908029
Number of successful extensions: 75814
Number of sequences better than 10.0: 187
Number of HSP's better than 10.0 without gapping: 58985
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 73347
length of database: 448,689,247
effective HSP length: 124
effective length of database: 275,931,827
effective search space used: 59325342805
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)