Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC008031A_C01 KMC008031A_c01
(606 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
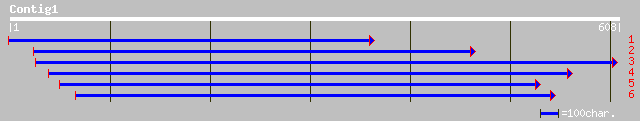
ref|NP_179803.1| unknown protein; protein id: At2g22125.1 [Arabi... 137 1e-31
ref|NP_177870.1| unknown protein; protein id: At1g77460.1 [Arabi... 83 2e-15
dbj|BAC42703.1| unknown protein [Arabidopsis thaliana] 83 2e-15
ref|NP_175078.1| hypothetical protein; protein id: At1g44120.1 [... 65 9e-10
gb|EAA21402.1| hypothetical protein [Plasmodium yoelii yoelii] 34 1.3
>ref|NP_179803.1| unknown protein; protein id: At2g22125.1 [Arabidopsis thaliana]
gi|20198149|gb|AAM15432.1| unknown protein [Arabidopsis
thaliana] gi|20198216|gb|AAM15466.1| unknown protein
[Arabidopsis thaliana]
Length = 109
Score = 137 bits (344), Expect = 1e-31
Identities = 68/74 (91%), Positives = 70/74 (93%)
Frame = -3
Query: 604 ESFSWSFESPPKGQKLQISCKNKSKVGKSKFGKVTIQIDRVVMLGAVAGEYTLLPASKSG 425
ESFSWSFESPPKGQKL ISCKNKSK+GKS FGKVTIQIDRVVMLGAVAGEY+LLP SKSG
Sbjct: 37 ESFSWSFESPPKGQKLHISCKNKSKMGKSSFGKVTIQIDRVVMLGAVAGEYSLLPESKSG 96
Query: 424 PPRNLEIEFQWSNK 383
PRNLEIEFQWSNK
Sbjct: 97 -PRNLEIEFQWSNK 109
>ref|NP_177870.1| unknown protein; protein id: At1g77460.1 [Arabidopsis thaliana]
gi|25406506|pir||H96803 unknown protein T5M16.5
[imported] - Arabidopsis thaliana
gi|12323397|gb|AAG51678.1|AC010704_22 unknown protein;
15069-22101 [Arabidopsis thaliana]
Length = 2110
Score = 83.2 bits (204), Expect = 2e-15
Identities = 41/78 (52%), Positives = 53/78 (67%), Gaps = 1/78 (1%)
Frame = -3
Query: 604 ESFSWSFESPPKGQKLQISCKNKSKVGKSKFGKVTIQIDRVVMLGAVAGEYTL-LPASKS 428
E F+W+F+ PPKGQKL I CK+KS GK+ G+VTIQID+VV G +G +L SK
Sbjct: 2030 EGFTWAFDVPPKGQKLHIICKSKSTFGKTTLGRVTIQIDKVVTEGEYSGSLSLNHENSKD 2089
Query: 427 GPPRNLEIEFQWSNKTND 374
R+L+IE WSN+T D
Sbjct: 2090 ASSRSLDIEIAWSNRTTD 2107
>dbj|BAC42703.1| unknown protein [Arabidopsis thaliana]
Length = 434
Score = 83.2 bits (204), Expect = 2e-15
Identities = 41/78 (52%), Positives = 53/78 (67%), Gaps = 1/78 (1%)
Frame = -3
Query: 604 ESFSWSFESPPKGQKLQISCKNKSKVGKSKFGKVTIQIDRVVMLGAVAGEYTL-LPASKS 428
E F+W+F+ PPKGQKL I CK+KS GK+ G+VTIQID+VV G +G +L SK
Sbjct: 354 EGFTWAFDVPPKGQKLHIICKSKSTFGKTTLGRVTIQIDKVVTEGEYSGSLSLNHENSKD 413
Query: 427 GPPRNLEIEFQWSNKTND 374
R+L+IE WSN+T D
Sbjct: 414 ASSRSLDIEIAWSNRTTD 431
>ref|NP_175078.1| hypothetical protein; protein id: At1g44120.1 [Arabidopsis thaliana]
gi|25405158|pir||E96505 hypothetical protein T7O23.25
[imported] - Arabidopsis thaliana
gi|12320824|gb|AAG50555.1|AC074228_10 hypothetical
protein [Arabidopsis thaliana]
Length = 2114
Score = 64.7 bits (156), Expect = 9e-10
Identities = 33/77 (42%), Positives = 48/77 (61%), Gaps = 2/77 (2%)
Frame = -3
Query: 604 ESFSWSFESPPKGQKLQISCKNKSKVGKSKFGKVTIQIDRVVMLGAVAGEYTLLPASK-- 431
ESF+W F +PP+GQ L+I CK+ + GKV I ID+V+ G+ +G + L SK
Sbjct: 2037 ESFTWDFAAPPRGQFLEIVCKSNNIFRNKNLGKVRIPIDKVLSEGSYSGIFKLNDESKKD 2096
Query: 430 SGPPRNLEIEFQWSNKT 380
+ R+LEIE WSN++
Sbjct: 2097 NSSDRSLEIEIVWSNQS 2113
>gb|EAA21402.1| hypothetical protein [Plasmodium yoelii yoelii]
Length = 707
Score = 34.3 bits (77), Expect = 1.3
Identities = 30/118 (25%), Positives = 53/118 (44%), Gaps = 2/118 (1%)
Frame = +3
Query: 3 CRRKKLIRNIICKHTTIFYLMVILHSKSQIQSCTCTVPKAYIEQTRYFTLN*RHAKNQTT 182
CR+ +++ IC IFY + IL+ ++ I SC + KN+T
Sbjct: 6 CRKHNILKRGICFFLYIFYYLNILNFENIIISC-------------------YNVKNRTE 46
Query: 183 DTRNQNKPGLSAPSVYPRK*RNKLQKNMMTKE--SVWNYRPKKKTAIYT*YLFFQKSK 350
+N+NK S++ +N + N+ + ++ NY +K+T T FF K+K
Sbjct: 47 FYKNKNKNXKKCVSLFIDNTKN-VATNIRKRNAFNLLNYAKEKRTKRKTELNFFSKNK 103
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 513,408,717
Number of Sequences: 1393205
Number of extensions: 11025154
Number of successful extensions: 25297
Number of sequences better than 10.0: 12
Number of HSP's better than 10.0 without gapping: 24609
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 25288
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 23997478008
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)