Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC007559A_C01 KMC007559A_c01
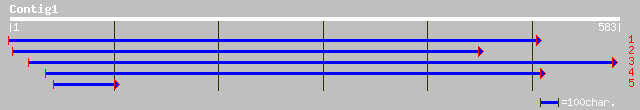
(577 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|XP_085200.1| hypothetical protein XP_085200 [Homo sapiens] 34 1.2
ref|NP_241969.1| BH1103~unknown conserved protein [Bacillus halo... 34 1.2
ref|XP_286971.1| hypothetical protein XP_286971 [Mus musculus] 33 2.0
ref|NP_809392.1| putative glycosyltransferase [Bacteroides theta... 33 2.0
gb|EAA06230.1| agCP13655 [Anopheles gambiae str. PEST] 33 3.4
>ref|XP_085200.1| hypothetical protein XP_085200 [Homo sapiens]
Length = 104
Score = 34.3 bits (77), Expect = 1.2
Identities = 18/50 (36%), Positives = 24/50 (48%)
Frame = +1
Query: 133 RW*IHGPWPP*PRAGVTSLSWLPPIPQSLILPTSPLDPL*LCSAHLLCLW 282
RW +H PWPP +T + WL P P S+ L + CS L +W
Sbjct: 11 RWLVHWPWPP---RAMTLIHWLHPPPCSMALGVT-------CSCPLPSVW 50
>ref|NP_241969.1| BH1103~unknown conserved protein [Bacillus halodurans]
gi|25489661|pir||G83787 hypothetical protein BH1103
[imported] - Bacillus halodurans (strain C-125)
gi|10173718|dbj|BAB04822.1| BH1103~unknown conserved
protein [Bacillus halodurans]
Length = 368
Score = 34.3 bits (77), Expect = 1.2
Identities = 18/36 (50%), Positives = 21/36 (58%), Gaps = 3/36 (8%)
Frame = -3
Query: 176 PARGYGGHGPWIHHLLLC---FIFSFSLLYMYFLFP 78
P GY G+G W+ LL FIF SLLY+Y FP
Sbjct: 35 PLIGYAGNGAWLSILLSAVTGFIFVASLLYLYRKFP 70
>ref|XP_286971.1| hypothetical protein XP_286971 [Mus musculus]
Length = 113
Score = 33.5 bits (75), Expect = 2.0
Identities = 32/104 (30%), Positives = 50/104 (47%), Gaps = 5/104 (4%)
Frame = +3
Query: 156 ASIASSRCHKSLMVATNTSKLNPSNLTLGSLIIMFRPFAMFMGGKCEE-----IPNSVIF 320
+SIAS R K A N + S+L++ S I++FR +C E IP+ VIF
Sbjct: 3 SSIASCRLAK----AGNLPMASFSSLSVSSFIMVFRSCCHCHCRECWEVSVTLIPDQVIF 58
Query: 321 SSLYISAWAVTLDKRVKTHSTSCCFIIGIKCSSLSEQGPAKIVP 452
+SL +S+ ++ + + + S+S SS S P I P
Sbjct: 59 ASLSMSSSSLLISSSISSSSSSAS--SSSSSSSSSSSCPPGIFP 100
>ref|NP_809392.1| putative glycosyltransferase [Bacteroides thetaiotaomicron
VPI-5482] gi|29337782|gb|AAO75586.1| putative
glycosyltransferase [Bacteroides thetaiotaomicron
VPI-5482]
Length = 274
Score = 33.5 bits (75), Expect = 2.0
Identities = 19/60 (31%), Positives = 29/60 (47%)
Frame = +3
Query: 108 KGKDETQEKVVNPRSMASIASSRCHKSLMVATNTSKLNPSNLTLGSLIIMFRPFAMFMGG 287
KG D+T K+VN K L +T+K+ P + +G+ + M R F F+GG
Sbjct: 146 KGNDQTTSKIVN-------------KYLCKNMDTTKIQPVDWAIGAALFMRRDFYAFLGG 192
>gb|EAA06230.1| agCP13655 [Anopheles gambiae str. PEST]
Length = 1955
Score = 32.7 bits (73), Expect = 3.4
Identities = 15/50 (30%), Positives = 30/50 (60%)
Frame = -1
Query: 481 DFLAEEICVFGTIFAGPCSLRELHFMPMMKQQLVE*VFTRLSSVTAHALI 332
D A+ + V G++F G C +R LH + +++Q+++E V ++ + A I
Sbjct: 148 DSNAKNLIVCGSLFQGICHIRNLHNISIVEQEVLEAVVANTANASTVAFI 197
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 514,987,276
Number of Sequences: 1393205
Number of extensions: 11342476
Number of successful extensions: 31180
Number of sequences better than 10.0: 13
Number of HSP's better than 10.0 without gapping: 30079
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 31160
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 21530810025
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)