Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC007216A_C01 KMC007216A_c01
(1388 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
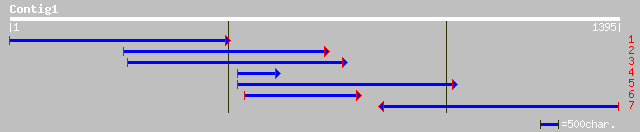
ref|NP_524664.1| spaghetti CG13570-PA gi|7242524|emb|CAB64598.2|... 45 0.003
gb|AAO24976.1| RE03224p [Drosophila melanogaster] 45 0.003
gb|EAA09485.1| agCP15078 [Anopheles gambiae str. PEST] 37 0.58
ref|XP_235372.1| similar to sperm associated antigen 1; TPR-cont... 37 0.75
ref|NP_798522.1| hypothetical protein [Vibrio parahaemolyticus R... 37 0.98
>ref|NP_524664.1| spaghetti CG13570-PA gi|7242524|emb|CAB64598.2| spaghetti [Drosophila
melanogaster] gi|7291754|gb|AAF47175.1| CG13570-PA
[Drosophila melanogaster]
Length = 534
Score = 45.1 bits (105), Expect = 0.003
Identities = 38/155 (24%), Positives = 69/155 (44%), Gaps = 4/155 (2%)
Frame = -3
Query: 1083 RKPEIKASVQELASRAASRALAEAA---KNITP-PTTAYQFEVSWKGFSGDLVLQTRLLK 916
R PE+ E +A+S + + + P PT+ QF V+WK SG Q LK
Sbjct: 382 RSPEVNTVQTEKIEQASSNNAMSPSPIERFLPPAPTSTAQFHVTWKELSGPQKYQ--YLK 439
Query: 915 ATSPPELPKILKNALSSTLLIEIIKCVASFFTEDMDLAVSFMENLDPKFPRFDMIAMCLP 736
+ P L KIL S ++++ + FF + + + + K F ++AM +
Sbjct: 440 SIEVPNLCKILGAGFDSDTFADLLRTIHDFFVPNKEPNTAAVLLEISKNDEFTILAMLMS 499
Query: 735 SADKNDLCKIWDEVFCNEATPMEYAEILDNLRSKF 631
+ +K + I + + + P + +LDNL ++
Sbjct: 500 AEEKKMVSSILNAI---KNWPSKNPAVLDNLFKEY 531
>gb|AAO24976.1| RE03224p [Drosophila melanogaster]
Length = 534
Score = 45.1 bits (105), Expect = 0.003
Identities = 38/155 (24%), Positives = 69/155 (44%), Gaps = 4/155 (2%)
Frame = -3
Query: 1083 RKPEIKASVQELASRAASRALAEAA---KNITP-PTTAYQFEVSWKGFSGDLVLQTRLLK 916
R PE+ E +A+S + + + P PT+ QF V+WK SG Q LK
Sbjct: 382 RSPEVNTVQTEKIEQASSNNAMSPSPIERFLPPAPTSTAQFHVTWKELSGPQKYQ--YLK 439
Query: 915 ATSPPELPKILKNALSSTLLIEIIKCVASFFTEDMDLAVSFMENLDPKFPRFDMIAMCLP 736
+ P L KIL S ++++ + FF + + + + K F ++AM +
Sbjct: 440 SIEVPNLCKILGAGFDSDTFADLLRTIHDFFVPNKEPNTAAVLLEISKNDEFTILAMLMS 499
Query: 735 SADKNDLCKIWDEVFCNEATPMEYAEILDNLRSKF 631
+ +K + I + + + P + +LDNL ++
Sbjct: 500 AEEKKMVSSILNAI---KNWPSKNPAVLDNLFKEY 531
>gb|EAA09485.1| agCP15078 [Anopheles gambiae str. PEST]
Length = 379
Score = 37.4 bits (85), Expect = 0.58
Identities = 32/110 (29%), Positives = 50/110 (45%), Gaps = 5/110 (4%)
Frame = -3
Query: 1323 SETKVNGGSIPSISHVTQKSGPAEVHQTKGNERQVPAKESLLMEIDNRDTRARNSTQGQG 1144
S T GG + H Q PA K + VPA ++ + I+N+ + +
Sbjct: 137 SGTAAAGGGV----HQKQSDAPA----AKTLQPPVPASKASGLTIENKRSLKSELLESDV 188
Query: 1143 NGSKEGSTSSNSLEQRNHRARKPE-----IKASVQELASRAASRALAEAA 1009
+GS+EGSTS S +++ R R + I A +L + A+ A A AA
Sbjct: 189 SGSREGSTSPTSRKRKKVRRRSVDTNNLLIAAETLKLQKQQAAAAAAAAA 238
>ref|XP_235372.1| similar to sperm associated antigen 1; TPR-containing protein
involved in spermatogenesis [Mus musculus] [Rattus
norvegicus]
Length = 844
Score = 37.0 bits (84), Expect = 0.75
Identities = 45/194 (23%), Positives = 85/194 (43%), Gaps = 3/194 (1%)
Frame = -3
Query: 1197 MEIDNRDTRARNSTQGQGNGSKEGSTSSNSLEQRNHRARKPEIKASVQELASRAASRALA 1018
++ID+++ +A + G + +S E +P ++V+E +A RA
Sbjct: 665 LQIDSKNVKASYRLELAQKGLEVDDSSDEEPE-------RPAEASAVEE--GWSAERA-- 713
Query: 1017 EAAKNITPPTTAYQFEVSWKGFSG--DLVLQTRLLKATSPPELPKILKNALSSTLLIEII 844
+ P AY+F S D +LL T+P +LP +L N L +L+ I+
Sbjct: 714 -GKIEVCKPRNAYEFGQVLSTISARKDEEACAQLLVFTAPQDLPVLLSNKLEGDMLLLIM 772
Query: 843 KCVAS-FFTEDMDLAVSFMENLDPKFPRFDMIAMCLPSADKNDLCKIWDEVFCNEATPME 667
+ + S +D L + L K RF+M+ K + +++D++ +A +
Sbjct: 773 QSLKSHLVAKDPSLVCKHLLYLS-KAERFEMMLALTSKDQKEQMAQLFDDLSDAQADCLT 831
Query: 666 YAEILDNLRSKFYL 625
AE + LR ++ L
Sbjct: 832 -AEDIQALRRQYVL 844
>ref|NP_798522.1| hypothetical protein [Vibrio parahaemolyticus RIMD 2210633]
gi|28807136|dbj|BAC60406.1| hypothetical protein [Vibrio
parahaemolyticus]
Length = 613
Score = 36.6 bits (83), Expect = 0.98
Identities = 29/91 (31%), Positives = 41/91 (44%), Gaps = 1/91 (1%)
Frame = -3
Query: 1284 SHVTQKSGPAEVHQTKGNERQVPAKESLLMEIDNRDTRARNSTQGQGNGSKEGSTSSN-S 1108
S V + P E+ +K N + V S L + + + T G G KEGSTS N S
Sbjct: 109 SKVIKNFAPLEIFASKLNAKNVEDMLSYLATLGDEQLHSLFQTTKHGAGFKEGSTSPNGS 168
Query: 1107 LEQRNHRARKPEIKASVQELASRAASRALAE 1015
LE+ H K + Q L S+ ++ L E
Sbjct: 169 LERLEHTIDK--LSFLFQSLFSKPITKLLPE 197
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,154,323,733
Number of Sequences: 1393205
Number of extensions: 25072915
Number of successful extensions: 76074
Number of sequences better than 10.0: 34
Number of HSP's better than 10.0 without gapping: 71542
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 75974
length of database: 448,689,247
effective HSP length: 127
effective length of database: 271,752,212
effective search space used: 91036991020
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)