Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC007151A_C01 KMC007151A_c01
(580 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
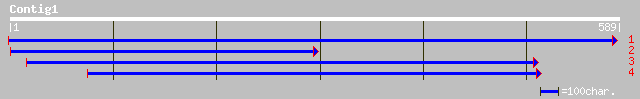
ref|NP_188161.1| hypothetical protein; protein id: At3g15420.1 [... 42 0.004
gb|AAO51261.1| similar to Dictyostelium discoideum (Slime mold).... 32 7.5
gb|EAA22091.1| unknown protein [Plasmodium yoelii yoelii] 32 7.5
>ref|NP_188161.1| hypothetical protein; protein id: At3g15420.1 [Arabidopsis
thaliana] gi|7021727|gb|AAF35408.1| hypothetical protein
[Arabidopsis thaliana] gi|15795107|dbj|BAB02371.1|
gene_id:MJK13.8~unknown protein [Arabidopsis thaliana]
Length = 107
Score = 42.4 bits (98), Expect = 0.004
Identities = 23/65 (35%), Positives = 38/65 (58%)
Frame = -2
Query: 447 IGEYEETIGTSIAFTEQDTRVVHEVTGPSEKNFFSGTRLIDSRQPSTKQVRPLCQLHKVL 268
+GEY ETIGT +AF+E++ EV+ +++ P K + P+ +LHK+L
Sbjct: 46 VGEYIETIGTCLAFSEKE-----EVSAS------------ENQMPHKKIIEPVAKLHKIL 88
Query: 267 KFKLS 253
KF+L+
Sbjct: 89 KFRLA 93
>gb|AAO51261.1| similar to Dictyostelium discoideum (Slime mold). Prespore-specific
protein (Fragment)
Length = 462
Score = 31.6 bits (70), Expect = 7.5
Identities = 20/65 (30%), Positives = 29/65 (43%)
Frame = +2
Query: 386 TRVSCSVKAILVPIVSSYSPICRQQQTIINLTRPSSIQRMEKQN*SNLNPHLKNSNYQWK 565
+R S + VP S+ SPI Q T+ + +S+ N SN N + N+N
Sbjct: 233 SRSSSPLSLSCVPSPSNSSPIASPQLTVPSTPLLTSVNGSRNNNNSNNNSNNNNNNQIQN 292
Query: 566 KNNEN 580
NN N
Sbjct: 293 NNNNN 297
>gb|EAA22091.1| unknown protein [Plasmodium yoelii yoelii]
Length = 1014
Score = 31.6 bits (70), Expect = 7.5
Identities = 17/48 (35%), Positives = 27/48 (55%), Gaps = 1/48 (2%)
Frame = -3
Query: 326 IQGNLQQSK*DHCVSFTRFSN-LNYHLILKFEFPQLRKRPNEFET*YM 186
I N+ +SK +H + TR + Y L+ K +FP + K N+ +T YM
Sbjct: 610 IHENIHESKNNHYNNITRVRRKVEYKLLFKKKFPFILKHKNKIQTFYM 657
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 483,200,919
Number of Sequences: 1393205
Number of extensions: 10006223
Number of successful extensions: 22802
Number of sequences better than 10.0: 6
Number of HSP's better than 10.0 without gapping: 21778
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 22734
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 21426319650
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)