Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC007108A_C01 KMC007108A_c01
(572 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
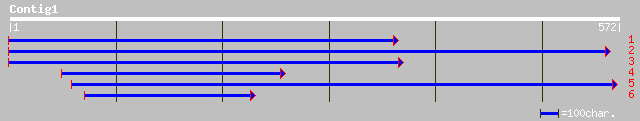
ref|NP_191595.1| putative protein; protein id: At3g60360.1, supp... 96 4e-19
ref|NP_050111.1| hypothetical protein [Lactobacillus bacteriopha... 34 1.1
ref|NP_037313.1| protein kinase, cAMP dependent, regulatory, typ... 34 1.1
dbj|BAB89638.1| P0468B07.17 [Oryza sativa (japonica cultivar-gro... 34 1.1
ref|NP_002725.1| protein kinase, cAMP-dependent, regulatory, typ... 34 1.5
>ref|NP_191595.1| putative protein; protein id: At3g60360.1, supported by cDNA:
13387., supported by cDNA: gi_14532503, supported by
cDNA: gi_20855864 [Arabidopsis thaliana]
gi|11281657|pir||T47847 hypothetical protein T8B10.20 -
Arabidopsis thaliana gi|7287984|emb|CAB81822.1| putative
protein [Arabidopsis thaliana]
gi|14532504|gb|AAK63980.1| AT3g60360/T8B10_20
[Arabidopsis thaliana] gi|20855865|gb|AAM26635.1|
AT3g60360/T8B10_20 [Arabidopsis thaliana]
gi|21537368|gb|AAM61709.1| unknown [Arabidopsis
thaliana] gi|23397168|gb|AAN31867.1| unknown protein
[Arabidopsis thaliana]
Length = 228
Score = 95.5 bits (236), Expect = 4e-19
Identities = 42/70 (60%), Positives = 59/70 (84%)
Frame = -2
Query: 571 IHTNIKRKIDRSYKELEARRTRLSQLQKMYMDMSMKKELQKNGRKRQLREDEIVSPTPEP 392
I +IK+K+DRSY++LE R++R L+K+Y DMSM+KELQK GRKR+LR+DE+++P +
Sbjct: 159 IPKDIKKKMDRSYRDLEGRKSRAKDLEKLYTDMSMQKELQKKGRKRKLRDDELLNPNGKA 218
Query: 391 VYKWRAERKR 362
VYKWRA+RKR
Sbjct: 219 VYKWRADRKR 228
>ref|NP_050111.1| hypothetical protein [Lactobacillus bacteriophage phi adh]
gi|5730260|emb|CAB52481.1| hypothetical protein
[Lactobacillus bacteriophage phi adh]
Length = 294
Score = 34.3 bits (77), Expect = 1.1
Identities = 16/40 (40%), Positives = 24/40 (60%)
Frame = -2
Query: 532 KELEARRTRLSQLQKMYMDMSMKKELQKNGRKRQLREDEI 413
KEL+ + +Q Y+D KKELQK K+Q R+++I
Sbjct: 59 KELQTSGKDFTNIQDQYIDDETKKELQKKQEKKQERQEKI 98
>ref|NP_037313.1| protein kinase, cAMP dependent, regulatory, type 1 [Rattus
norvegicus] gi|125196|sp|P09456|KAP0_RAT CAMP-DEPENDENT
PROTEIN KINASE TYPE I-ALPHA REGULATORY CHAIN
gi|279506|pir||OKRT1R protein kinase (EC 2.7.1.37),
cAMP-dependent, type I-alpha regulatory chain - rat
gi|206673|gb|AAB54276.1| R-I subunit
Length = 381
Score = 34.3 bits (77), Expect = 1.1
Identities = 26/72 (36%), Positives = 34/72 (47%), Gaps = 7/72 (9%)
Frame = -2
Query: 562 NIKRKIDRSYKEL-EARRTRLSQLQKMYMDMSMKKE------LQKNGRKRQLREDEIVSP 404
NI+ + S +L AR R + Y + K+E LQK+G + REDEI P
Sbjct: 26 NIQALLKDSIVQLCTARPERPMAFLREYFERLEKEEARQIQSLQKSGIRTDSREDEISPP 85
Query: 403 TPEPVYKWRAER 368
P PV K R R
Sbjct: 86 PPNPVVKGRRRR 97
>dbj|BAB89638.1| P0468B07.17 [Oryza sativa (japonica cultivar-group)]
Length = 214
Score = 34.3 bits (77), Expect = 1.1
Identities = 13/27 (48%), Positives = 23/27 (85%)
Frame = -2
Query: 532 KELEARRTRLSQLQKMYMDMSMKKELQ 452
+ELE R+ R+ +L+K+Y DM+++KEL+
Sbjct: 175 RELEERKQRVQKLEKLYADMALQKELK 201
>ref|NP_002725.1| protein kinase, cAMP-dependent, regulatory, type I, alpha;
tissue-specific extinguisher 1 [Homo sapiens]
gi|125193|sp|P10644|KAP0_HUMAN cAMP-dependent protein
kinase type I-alpha regulatory chain (Tissue-specific
extinguisher-1) (TSE1) gi|279505|pir||OKHU1R protein
kinase (EC 2.7.1.37), cAMP-dependent, type I-alpha
regulatory chain - human gi|179895|gb|AAB50921.1|
cAMP-dependent protein kinase type I-alpha subunit [Homo
sapiens] gi|179922|gb|AAB50922.1| cAMP-dependent protein
kinase regulatory subunit type 1 [Homo sapiens]
gi|23273780|gb|AAH36285.1| protein kinase,
cAMP-dependent, regulatory, type I, alpha (tissue
specific extinguisher 1) [Homo sapiens]
Length = 381
Score = 33.9 bits (76), Expect = 1.5
Identities = 26/72 (36%), Positives = 33/72 (45%), Gaps = 7/72 (9%)
Frame = -2
Query: 562 NIKRKIDRSYKEL-EARRTRLSQLQKMYMDMSMKKE------LQKNGRKRQLREDEIVSP 404
NI+ + S +L AR R + Y + K+E LQK G + REDEI P
Sbjct: 26 NIQALLKDSIVQLCTARPERPMAFLREYFERLEKEEAKQIQNLQKAGTRTDSREDEISPP 85
Query: 403 TPEPVYKWRAER 368
P PV K R R
Sbjct: 86 PPNPVVKGRRRR 97
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 436,377,139
Number of Sequences: 1393205
Number of extensions: 8324137
Number of successful extensions: 20913
Number of sequences better than 10.0: 31
Number of HSP's better than 10.0 without gapping: 20282
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 20900
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 21243732558
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)