Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC007004A_C01 KMC007004A_c01
(571 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
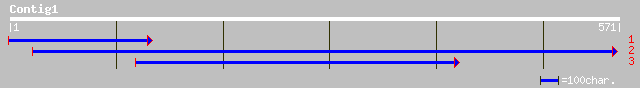
ref|NP_564467.1| acetyl-CoA carboxylase, putative; protein id: A... 101 6e-21
gb|AAG52317.1|AC021666_6 unknown protein; 24499-21911 [Arabidops... 101 6e-21
ref|NP_564162.1| expressed protein; protein id: At1g22200.1, sup... 96 3e-19
gb|AAF18633.1|AC006228_4 F5J5.4 [Arabidopsis thaliana] 88 7e-17
gb|AAH44474.1| Similar to serologically defined breast cancer an... 76 3e-13
>ref|NP_564467.1| acetyl-CoA carboxylase, putative; protein id: At1g36050.1
[Arabidopsis thaliana]
Length = 416
Score = 101 bits (252), Expect = 6e-21
Identities = 46/50 (92%), Positives = 50/50 (100%)
Frame = -3
Query: 569 FTEEHVSFLHFLTNVCAIVGGVFTVSGIVDSFIYHGQKAIKKKMELGKFS 420
FTEEH+SFLHFLTNVCAIVGGVFTVSGI+D+FIYHGQKAIKKKME+GKFS
Sbjct: 367 FTEEHISFLHFLTNVCAIVGGVFTVSGIIDAFIYHGQKAIKKKMEIGKFS 416
>gb|AAG52317.1|AC021666_6 unknown protein; 24499-21911 [Arabidopsis thaliana]
gi|27808598|gb|AAO24579.1| At1g36050 [Arabidopsis
thaliana]
Length = 386
Score = 101 bits (252), Expect = 6e-21
Identities = 46/50 (92%), Positives = 50/50 (100%)
Frame = -3
Query: 569 FTEEHVSFLHFLTNVCAIVGGVFTVSGIVDSFIYHGQKAIKKKMELGKFS 420
FTEEH+SFLHFLTNVCAIVGGVFTVSGI+D+FIYHGQKAIKKKME+GKFS
Sbjct: 337 FTEEHISFLHFLTNVCAIVGGVFTVSGIIDAFIYHGQKAIKKKMEIGKFS 386
>ref|NP_564162.1| expressed protein; protein id: At1g22200.1, supported by cDNA:
16380., supported by cDNA: gi_13878150, supported by
cDNA: gi_21281041 [Arabidopsis thaliana]
gi|25350066|pir||F86354 F16L1.7 protein - Arabidopsis
thaliana gi|9454530|gb|AAF87853.1|AC073942_7 Contains
similarity to a PR00989 protein from Homo sapiens
gi|7959731. EST gb|AI995648 comes from this gene.
[Arabidopsis thaliana]
gi|13878151|gb|AAK44153.1|AF370338_1 unknown protein
[Arabidopsis thaliana] gi|21281042|gb|AAM44956.1|
unknown protein [Arabidopsis thaliana]
gi|21553754|gb|AAM62847.1| unknown [Arabidopsis
thaliana]
Length = 386
Score = 95.9 bits (237), Expect = 3e-19
Identities = 43/50 (86%), Positives = 48/50 (96%)
Frame = -3
Query: 569 FTEEHVSFLHFLTNVCAIVGGVFTVSGIVDSFIYHGQKAIKKKMELGKFS 420
F E+HV FLHFLTNVCAIVGG+FTVSGIVDSFIYHGQ+AIKKKME+GKF+
Sbjct: 337 FEEQHVEFLHFLTNVCAIVGGIFTVSGIVDSFIYHGQRAIKKKMEIGKFN 386
>gb|AAF18633.1|AC006228_4 F5J5.4 [Arabidopsis thaliana]
Length = 440
Score = 88.2 bits (217), Expect = 7e-17
Identities = 46/74 (62%), Positives = 50/74 (67%), Gaps = 24/74 (32%)
Frame = -3
Query: 569 FTEEHVSFLHFLTNVCAIVGG------------------------VFTVSGIVDSFIYHG 462
FTEEH+SFLHFLTNVCAIVGG VFTVSGI+D+FIYHG
Sbjct: 367 FTEEHISFLHFLTNVCAIVGGISLISIYHNNTCWLTHIKIRNETCVFTVSGIIDAFIYHG 426
Query: 461 QKAIKKKMELGKFS 420
QKAIKKKME+GKFS
Sbjct: 427 QKAIKKKMEIGKFS 440
>gb|AAH44474.1| Similar to serologically defined breast cancer antigen 84 [Danio
rerio]
Length = 383
Score = 75.9 bits (185), Expect = 3e-13
Identities = 34/50 (68%), Positives = 42/50 (84%)
Frame = -3
Query: 569 FTEEHVSFLHFLTNVCAIVGGVFTVSGIVDSFIYHGQKAIKKKMELGKFS 420
FTE+ SF HFLT VCAI+GGVFTV+G++DS IYH +AI+KK+ELGK S
Sbjct: 334 FTEKQRSFTHFLTGVCAIIGGVFTVAGLIDSLIYHSARAIQKKIELGKAS 383
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 475,495,990
Number of Sequences: 1393205
Number of extensions: 9632312
Number of successful extensions: 20666
Number of sequences better than 10.0: 47
Number of HSP's better than 10.0 without gapping: 20082
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 20663
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 20956655091
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)