Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC006879A_C01 KMC006879A_c01
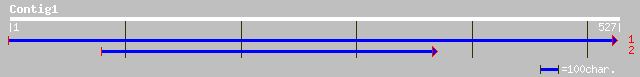
(527 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|EAA06809.1| agCP7316 [Anopheles gambiae str. PEST] 35 0.42
gb|AAO51651.1| similar to Dictyostelium discoideum (Slime mold).... 33 2.1
gb|AAO51816.1| similar to Dictyostelium discoideum (Slime mold).... 33 2.7
ref|ZP_00086147.1| hypothetical protein [Pseudomonas fluorescens... 32 3.5
ref|NP_014410.1| Low-affinity phosphate transporter; Pho91p [Sac... 32 4.6
>gb|EAA06809.1| agCP7316 [Anopheles gambiae str. PEST]
Length = 157
Score = 35.4 bits (80), Expect = 0.42
Identities = 15/33 (45%), Positives = 20/33 (60%)
Frame = -1
Query: 215 SHSRCETFQFFLGLYSRWSESWGNRLCHKKNCS 117
S + CE+ QF+L +S W S G LCH K C+
Sbjct: 86 SPADCESLQFYLNPFSTWYSSNGLMLCHPKCCA 118
>gb|AAO51651.1| similar to Dictyostelium discoideum (Slime mold). Class VII
unconventional myosin
Length = 3446
Score = 33.1 bits (74), Expect = 2.1
Identities = 18/63 (28%), Positives = 31/63 (48%)
Frame = +2
Query: 26 NHEIKDSRSANSNLEIKILPPSPSHPHLA*SYNSFCDRDDFPNSHSTSSKVRGKIGKSHN 205
N+ + N+N I+ SPS P + NS + ++ N+ ++ S G IGK+H
Sbjct: 300 NNNFSSIQGTNTNTNTNIISSSPSSPLTSMVSNSSNNNNNNNNNSTSQSNNGGVIGKTHQ 359
Query: 206 VSV 214
S+
Sbjct: 360 KSI 362
>gb|AAO51816.1| similar to Dictyostelium discoideum (Slime mold). Hypothetical 97.7
kDa protein
Length = 565
Score = 32.7 bits (73), Expect = 2.7
Identities = 19/49 (38%), Positives = 28/49 (56%), Gaps = 1/49 (2%)
Frame = +2
Query: 23 LNHEI-KDSRSANSNLEIKILPPSPSHPHLA*SYNSFCDRDDFPNSHST 166
LN EI K S N NL +++LPP+ L ++N +RD+ P S +T
Sbjct: 466 LNLEILKFSSEFNQNLTLEMLPPNLKTLQLGSNFNKIINRDNMPESLTT 514
>ref|ZP_00086147.1| hypothetical protein [Pseudomonas fluorescens PfO-1]
Length = 240
Score = 32.3 bits (72), Expect = 3.5
Identities = 31/117 (26%), Positives = 47/117 (39%), Gaps = 13/117 (11%)
Frame = -2
Query: 409 SNHIQLQPFKKIAFTFWRDPTFQGKINVLSL*TKHALN**YVTDKLY*GGGF-------M 251
+NH L+ F +F +RDP QG ++L + D++ G GF M
Sbjct: 13 ANHGWLKSFHTFSFASYRDPREQGFSDLLVI----------NDDRVAAGKGFGQHPHRDM 62
Query: 250 NIFHYFLDGSLTHT------HVVRLSNFSSDFTRGGVRVGEIVSVTKRIVALSQMWM 98
IF Y L+G+L H V+R + GV E + V Q+W+
Sbjct: 63 EIFSYVLEGALEHKDTLGTGSVIRPGDVQLMSAGSGVAHSEFNHSATKPVHFLQIWI 119
>ref|NP_014410.1| Low-affinity phosphate transporter; Pho91p [Saccharomyces
cerevisiae] gi|732208|sp|P27514|YN86_YEAST Hypothetical
99.5 kDa protein in URK1-SMM1 intergenic region
gi|626467|pir||S45135 probable membrane protein YNR013c
- yeast (Saccharomyces cerevisiae)
gi|496729|emb|CAA54581.1| N2052 [Saccharomyces
cerevisiae] gi|1302492|emb|CAA96290.1| ORF YNR013c
[Saccharomyces cerevisiae]
Length = 894
Score = 32.0 bits (71), Expect = 4.6
Identities = 13/24 (54%), Positives = 17/24 (70%)
Frame = -1
Query: 431 IGYLAFNIQPYSTPTLQKNCFHIL 360
I L FN+ P++ +LQKNCF IL
Sbjct: 440 IALLTFNLTPFTQDSLQKNCFAIL 463
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 446,889,845
Number of Sequences: 1393205
Number of extensions: 9396324
Number of successful extensions: 18453
Number of sequences better than 10.0: 13
Number of HSP's better than 10.0 without gapping: 17982
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 18450
length of database: 448,689,247
effective HSP length: 115
effective length of database: 288,470,672
effective search space used: 17308240320
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)