Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC006515A_C01 KMC006515A_c01
(526 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
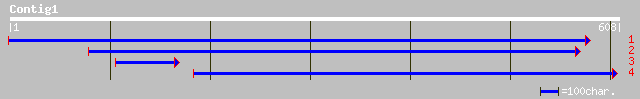
ref|NP_568458.1| putative protein; protein id: At5g24810.1, supp... 173 1e-42
ref|NP_702031.1| hypothetical protein [Plasmodium falciparum 3D7... 72 5e-12
gb|EAA16198.1| hypothetical protein [Plasmodium yoelii yoelii] 56 2e-07
ref|NP_595922.1| hypothetical protein [Schizosaccharomyces pombe... 54 1e-06
ref|NP_343936.1| ABC transporter, ABC1 family, putative [Sulfolo... 53 2e-06
>ref|NP_568458.1| putative protein; protein id: At5g24810.1, supported by cDNA:
gi_16649082 [Arabidopsis thaliana]
gi|16649083|gb|AAL24393.1| Unknown protein [Arabidopsis
thaliana]
Length = 1009
Score = 173 bits (439), Expect = 1e-42
Identities = 81/104 (77%), Positives = 94/104 (89%)
Frame = +3
Query: 183 VRMGWGNIYRRRIRVFTMALAIYLDYKGVQQREKWTNKSRQPVLWEKAHERNAKRVLNLI 362
V MG GNIYRRR++VF++A+ IYLDYKGVQQ+EKW KS+ P LW+KAH+RNAKRVLNLI
Sbjct: 46 VSMGLGNIYRRRMKVFSIAILIYLDYKGVQQKEKWIKKSKVPALWDKAHDRNAKRVLNLI 105
Query: 363 LEMEGLWVKLGQYMSTRADVLPAAYIRLLKQLQDSLPPRPLEEV 494
+E+EGLWVKLGQY+STRADVLP AYI LL QLQDSLPPRPL+EV
Sbjct: 106 VELEGLWVKLGQYLSTRADVLPQAYISLLTQLQDSLPPRPLQEV 149
>ref|NP_702031.1| hypothetical protein [Plasmodium falciparum 3D7]
gi|23497208|gb|AAN36755.1|AE014818_20 hypothetical
protein [Plasmodium falciparum 3D7]
Length = 2770
Score = 71.6 bits (174), Expect = 5e-12
Identities = 34/103 (33%), Positives = 58/103 (56%), Gaps = 7/103 (6%)
Frame = +3
Query: 207 YRRRIRVFTMALAIYLDYKGVQQREKWTNKSRQPVLWEKAHERNAKRVLNLILEMEGL-- 380
+ RRIR ++Y++YK ++ K ++ WEK HE A ++LN I E+ G
Sbjct: 7 WNRRIRTIWYCSSLYVEYKNALRKSKKMPVEKKNEYWEKKHEEFATKMLNNIYELRGKEY 66
Query: 381 -----WVKLGQYMSTRADVLPAAYIRLLKQLQDSLPPRPLEEV 494
WVK+GQ++ST+ +++P AYI +LQD +P +++
Sbjct: 67 YYKSWWVKVGQFLSTQENIMPVAYIEKFTKLQDMMPTSSFDKI 109
>gb|EAA16198.1| hypothetical protein [Plasmodium yoelii yoelii]
Length = 76
Score = 56.2 bits (134), Expect = 2e-07
Identities = 22/53 (41%), Positives = 38/53 (71%)
Frame = +3
Query: 348 VLNLILEMEGLWVKLGQYMSTRADVLPAAYIRLLKQLQDSLPPRPLEEVILLM 506
+LN I E+ G WVK+GQ++ST+ +++P AYI +LQD +P P E++ +++
Sbjct: 1 MLNNIYELRGWWVKVGQFLSTQENIMPVAYIEKFTKLQDMMPTSPFEKIEIIL 53
>ref|NP_595922.1| hypothetical protein [Schizosaccharomyces pombe]
gi|7491371|pir||T39472 hypothetical protein SPBC15C4.02
- fission yeast (Schizosaccharomyces pombe)
gi|3116145|emb|CAA18893.1| hypothetical protein
[Schizosaccharomyces pombe]
Length = 594
Score = 53.5 bits (127), Expect = 1e-06
Identities = 40/135 (29%), Positives = 65/135 (47%), Gaps = 7/135 (5%)
Frame = +3
Query: 111 DHLLPRDHRSPAT---AFLLDQALRL----PVRMGWGNIYRRRIRVFTMALAIYLDYKGV 269
DH+ R+H PA + L ++ L PV+ G +R RV + DYK V
Sbjct: 67 DHIQKRNHHKPAVVGASITLMASVALVDFDPVKHA-GVSSKRAYRVVLAGFLCFSDYKKV 125
Query: 270 QQREKWTNKSRQPVLWEKAHERNAKRVLNLILEMEGLWVKLGQYMSTRADVLPAAYIRLL 449
+ + RQ L E H R A+R L + E G+++K+GQ++S V+P + +
Sbjct: 126 LGSSYASEEERQLALSE-CHLRCAERSLKVFEENGGIYIKIGQHLSAMGYVIPKEWTNTM 184
Query: 450 KQLQDSLPPRPLEEV 494
+LQD P L+++
Sbjct: 185 VKLQDRCPSTSLKDI 199
>ref|NP_343936.1| ABC transporter, ABC1 family, putative [Sulfolobus solfataricus]
gi|25393625|pir||G90433 ABC transporter, ABC1 family,
probable SSO2605 [imported] - Sulfolobus solfataricus
gi|13815908|gb|AAK42726.1| ABC transporter, ABC1 family,
putative [Sulfolobus solfataricus]
Length = 496
Score = 53.1 bits (126), Expect = 2e-06
Identities = 30/96 (31%), Positives = 52/96 (53%), Gaps = 1/96 (1%)
Frame = +3
Query: 210 RRRIRVFTMALAIYLDYKGVQQREKWTNKSRQPV-LWEKAHERNAKRVLNLILEMEGLWV 386
RR + VF L Y+ ++ NK + + + EK AK+ ++ ++E+ ++
Sbjct: 3 RRLLEVFFKLAPRVLAYR------EFRNKILKEIPIDEKEMAEEAKKFVDTLIELGPTFI 56
Query: 387 KLGQYMSTRADVLPAAYIRLLKQLQDSLPPRPLEEV 494
K GQ +S R D++P YI+ L +LQD +PP P +V
Sbjct: 57 KFGQILSVRPDIMPETYIKELARLQDEVPPAPFSQV 92
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 488,088,544
Number of Sequences: 1393205
Number of extensions: 11277963
Number of successful extensions: 40923
Number of sequences better than 10.0: 187
Number of HSP's better than 10.0 without gapping: 38196
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 40846
length of database: 448,689,247
effective HSP length: 115
effective length of database: 288,470,672
effective search space used: 17019769648
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)