Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC006359A_C01 KMC006359A_c01
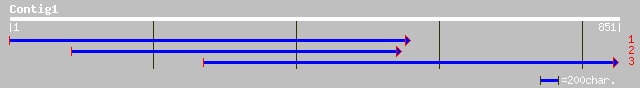
(755 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_565749.1| expressed protein; protein id: At2g32700.1, sup... 174 2e-42
ref|NP_567896.1| putative protein; protein id: At4g32551.1, supp... 159 5e-38
gb|AAG32022.1|AF277458_1 LEUNIG [Arabidopsis thaliana] 159 5e-38
pir||T08588 hypothetical protein L23H3.30 - Arabidopsis thaliana... 159 5e-38
dbj|BAB84838.1| putative flower development regulator LEUNI prot... 97 3e-19
>ref|NP_565749.1| expressed protein; protein id: At2g32700.1, supported by cDNA:
gi_13605814 [Arabidopsis thaliana]
gi|7486097|pir||T00798 hypothetical protein At2g32700
[imported] - Arabidopsis thaliana
gi|2914703|gb|AAC04493.1| expressed protein [Arabidopsis
thaliana] gi|13605815|gb|AAK32893.1|AF367306_1
At2g32700/F24L7.16 [Arabidopsis thaliana]
gi|25090107|gb|AAN72230.1| At2g32700/F24L7.16
[Arabidopsis thaliana]
Length = 787
Score = 174 bits (440), Expect = 2e-42
Identities = 75/105 (71%), Positives = 92/105 (87%)
Frame = -2
Query: 754 ICWDTNGDFLASLSQDSVKVWSLATGDCIHELNSSGNMFHSCVFHPSYSTLLVVGGYQSL 575
+CW NG+ +AS+S+D+VK+WSL++GDCIHEL++SGN FHS VFHPSY LLV+GGYQ++
Sbjct: 683 VCWSPNGELVASVSEDAVKLWSLSSGDCIHELSNSGNKFHSVVFHPSYPDLLVIGGYQAI 742
Query: 574 ELWNMAENKCMTIPAHECVISSLAQSPLTGMVASTSHDKSVKIWK 440
ELWN ENKCMT+ HECVIS+LAQSP TG+VAS SHDKSVKIWK
Sbjct: 743 ELWNTMENKCMTVAGHECVISALAQSPSTGVVASASHDKSVKIWK 787
Score = 35.0 bits (79), Expect = 1.2
Identities = 24/95 (25%), Positives = 45/95 (47%), Gaps = 1/95 (1%)
Frame = -2
Query: 724 ASLSQDSVKVWSLATGDCIHELNSSGNMFHSCVFHPSYSTLLVVGGYQSLELWNMAENKC 545
+S+ ++ K +S CI + S + C F L G + + +WNM +
Sbjct: 488 SSVHTETSKPFSFNEVSCIRK---SASKVICCSFSYDGKLLASAGHDKKVFIWNMETLQV 544
Query: 544 MTIPA-HECVISSLAQSPLTGMVASTSHDKSVKIW 443
+ P H +I+ + P + +A++S DK++KIW
Sbjct: 545 ESTPEEHAHIITDVRFRPNSTQLATSSFDKTIKIW 579
>ref|NP_567896.1| putative protein; protein id: At4g32551.1, supported by cDNA:
gi_11141604 [Arabidopsis thaliana]
Length = 931
Score = 159 bits (401), Expect = 5e-38
Identities = 72/108 (66%), Positives = 90/108 (82%), Gaps = 3/108 (2%)
Frame = -2
Query: 754 ICWDTNGDFLASLSQDSVKVWSLATG---DCIHELNSSGNMFHSCVFHPSYSTLLVVGGY 584
+CWD +GDFLAS+S+D VKVW+L TG +C+HEL+ +GN F SCVFHP+Y +LLV+G Y
Sbjct: 824 VCWDPSGDFLASVSEDMVKVWTLGTGSEGECVHELSCNGNKFQSCVFHPAYPSLLVIGCY 883
Query: 583 QSLELWNMAENKCMTIPAHECVISSLAQSPLTGMVASTSHDKSVKIWK 440
QSLELWNM+ENK MT+PAHE +I+SLA S TG+VAS SHDK VK+WK
Sbjct: 884 QSLELWNMSENKTMTLPAHEGLITSLAVSTATGLVASASHDKLVKLWK 931
>gb|AAG32022.1|AF277458_1 LEUNIG [Arabidopsis thaliana]
Length = 931
Score = 159 bits (401), Expect = 5e-38
Identities = 72/108 (66%), Positives = 90/108 (82%), Gaps = 3/108 (2%)
Frame = -2
Query: 754 ICWDTNGDFLASLSQDSVKVWSLATG---DCIHELNSSGNMFHSCVFHPSYSTLLVVGGY 584
+CWD +GDFLAS+S+D VKVW+L TG +C+HEL+ +GN F SCVFHP+Y +LLV+G Y
Sbjct: 824 VCWDPSGDFLASVSEDMVKVWTLGTGSEGECVHELSCNGNKFQSCVFHPAYPSLLVIGCY 883
Query: 583 QSLELWNMAENKCMTIPAHECVISSLAQSPLTGMVASTSHDKSVKIWK 440
QSLELWNM+ENK MT+PAHE +I+SLA S TG+VAS SHDK VK+WK
Sbjct: 884 QSLELWNMSENKTMTLPAHEGLITSLAVSTATGLVASASHDKLVKLWK 931
>pir||T08588 hypothetical protein L23H3.30 - Arabidopsis thaliana
gi|4914452|emb|CAB43692.1| putative protein [Arabidopsis
thaliana] gi|7270159|emb|CAB79972.1| putative protein
[Arabidopsis thaliana]
Length = 930
Score = 159 bits (401), Expect = 5e-38
Identities = 72/108 (66%), Positives = 90/108 (82%), Gaps = 3/108 (2%)
Frame = -2
Query: 754 ICWDTNGDFLASLSQDSVKVWSLATG---DCIHELNSSGNMFHSCVFHPSYSTLLVVGGY 584
+CWD +GDFLAS+S+D VKVW+L TG +C+HEL+ +GN F SCVFHP+Y +LLV+G Y
Sbjct: 823 VCWDPSGDFLASVSEDMVKVWTLGTGSEGECVHELSCNGNKFQSCVFHPAYPSLLVIGCY 882
Query: 583 QSLELWNMAENKCMTIPAHECVISSLAQSPLTGMVASTSHDKSVKIWK 440
QSLELWNM+ENK MT+PAHE +I+SLA S TG+VAS SHDK VK+WK
Sbjct: 883 QSLELWNMSENKTMTLPAHEGLITSLAVSTATGLVASASHDKLVKLWK 930
>dbj|BAB84838.1| putative flower development regulator LEUNI protein [Oryza sativa
(japonica cultivar-group)] gi|20804798|dbj|BAB92482.1|
putative flower development regulator, LEUNI protein
[Oryza sativa (japonica cultivar-group)]
Length = 883
Score = 96.7 bits (239), Expect = 3e-19
Identities = 50/98 (51%), Positives = 68/98 (69%), Gaps = 4/98 (4%)
Frame = -2
Query: 754 ICWDTNGDFLASLSQDSVKVWSLA---TGDCIHELNSSGNMFHSCVFHPSYSTLLVVGGY 584
+CWD+ GD LAS+S+DSV++WS A G+ ++EL+ SGN F SCVFHPSY LL
Sbjct: 782 LCWDSTGDNLASVSEDSVRIWSFAPGHDGEFVNELSCSGNKFQSCVFHPSYPYLL----- 836
Query: 583 QSLELWNMAENKCMTI-PAHECVISSLAQSPLTGMVAS 473
SLELW++ E MT+ AH+ ++++LA S TG VAS
Sbjct: 837 -SLELWDIREKNAMTVHSAHDGLVAALAASSATGKVAS 873
Score = 33.5 bits (75), Expect = 3.4
Identities = 21/78 (26%), Positives = 39/78 (49%), Gaps = 3/78 (3%)
Frame = -2
Query: 634 SCVFHPSYSTLLVVGGY-QSLELW--NMAENKCMTIPAHECVISSLAQSPLTGMVASTSH 464
+C S LL GG+ + + LW A ++ H +I+ + SP +A++S
Sbjct: 608 TCCHFSSDGKLLATGGHDKKVLLWCTEPALKPTSSLEEHSALITDVRFSPSMSRLATSSF 667
Query: 463 DKSVKIWK*N*LFWSYKT 410
DK+V++W + +S +T
Sbjct: 668 DKTVRVWDADNTSYSLRT 685
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 686,516,171
Number of Sequences: 1393205
Number of extensions: 15352357
Number of successful extensions: 41207
Number of sequences better than 10.0: 863
Number of HSP's better than 10.0 without gapping: 36789
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 40398
length of database: 448,689,247
effective HSP length: 120
effective length of database: 281,504,647
effective search space used: 36877108757
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)