Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
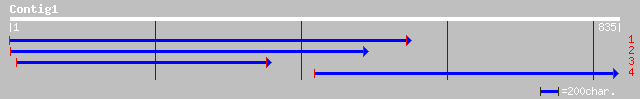
Query= KMC006334A_C01 KMC006334A_c01
(834 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_187074.1| putative O-linked GlcNAc transferase; protein i... 270 2e-71
dbj|BAB92672.1| putative O-linked GlcNAc transferase [Oryza sati... 208 7e-53
pir||E88499 protein K04G7.3 [imported] - Caenorhabditis elegans 108 7e-23
ref|NP_498563.1| O-linked N-acetylglucosamine transferase, nucle... 108 7e-23
gb|AAB63465.1| O-linked GlcNAc transferase [Caenorhabditis elegans] 108 7e-23
>ref|NP_187074.1| putative O-linked GlcNAc transferase; protein id: At3g04240.1,
supported by cDNA: gi_18139886, supported by cDNA:
gi_20259323 [Arabidopsis thaliana]
gi|6721161|gb|AAF26789.1|AC016829_13 putative O-linked
GlcNAc transferase [Arabidopsis thaliana]
gi|18139887|gb|AAL60196.1|AF441079_1 O-linked N-acetyl
glucosamine transferase [Arabidopsis thaliana]
gi|20259324|gb|AAM13988.1| putative O-linked GlcNAc
transferase [Arabidopsis thaliana]
gi|21436429|gb|AAM51415.1| putative O-linked GlcNAc
transferase [Arabidopsis thaliana]
Length = 977
Score = 270 bits (690), Expect = 2e-71
Identities = 125/159 (78%), Positives = 145/159 (90%)
Frame = +3
Query: 357 EVDEDLHLSLAHQMYKSGSYKKALEHSNIVYDRNPLRTDNLLLLGAIYYQLHDFDMCVAK 536
E D+D L+LAHQ+YK G +K+ALEHSN+VY RNPLRTDNLLL+GAIYYQL ++DMC+A+
Sbjct: 50 EGDDDARLALAHQLYKGGDFKQALEHSNMVYQRNPLRTDNLLLIGAIYYQLQEYDMCIAR 109
Query: 537 NEEALRVEPHFAECYGNMANALKEKGNIDLAIRYYLIAIEFRPNFADAWSNLASAYMRKG 716
NEEALR++P FAECYGNMANA KEKG+ D AIRYYLIAIE RPNFADAWSNLASAYMRKG
Sbjct: 110 NEEALRIQPQFAECYGNMANAWKEKGDTDRAIRYYLIAIELRPNFADAWSNLASAYMRKG 169
Query: 717 RLTEAAQCCRQALAINPLMVDAHSNLGNLMKAQGLVQEA 833
RL+EA QCC+QAL++NPL+VDAHSNLGNLMKAQGL+ EA
Sbjct: 170 RLSEATQCCQQALSLNPLLVDAHSNLGNLMKAQGLIHEA 208
Score = 84.7 bits (208), Expect = 1e-15
Identities = 49/141 (34%), Positives = 76/141 (53%)
Frame = +3
Query: 381 SLAHQMYKSGSYKKALEHSNIVYDRNPLRTDNLLLLGAIYYQLHDFDMCVAKNEEALRVE 560
+LA +SG +AL++ P D L LG +Y L + + AL++
Sbjct: 228 NLAGLFMESGDLNRALQYYKEAVKLKPAFPDAYLNLGNVYKALGRPTEAIMCYQHALQMR 287
Query: 561 PHFAECYGNMANALKEKGNIDLAIRYYLIAIEFRPNFADAWSNLASAYMRKGRLTEAAQC 740
P+ A +GN+A+ E+G +DLAIR+Y A+ P F +A++NL +A GR+ EA +C
Sbjct: 288 PNSAMAFGNIASIYYEQGQLDLAIRHYKQALSRDPRFLEAYNNLGNALKDIGRVDEAVRC 347
Query: 741 CRQALAINPLMVDAHSNLGNL 803
Q LA+ P A +NLGN+
Sbjct: 348 YNQCLALQPNHPQAMANLGNI 368
Score = 80.9 bits (198), Expect = 2e-14
Identities = 47/151 (31%), Positives = 74/151 (48%)
Frame = +3
Query: 381 SLAHQMYKSGSYKKALEHSNIVYDRNPLRTDNLLLLGAIYYQLHDFDMCVAKNEEALRVE 560
++A+ + G +A+ + I + P D L + Y + ++AL +
Sbjct: 126 NMANAWKEKGDTDRAIRYYLIAIELRPNFADAWSNLASAYMRKGRLSEATQCCQQALSLN 185
Query: 561 PHFAECYGNMANALKEKGNIDLAIRYYLIAIEFRPNFADAWSNLASAYMRKGRLTEAAQC 740
P + + N+ N +K +G I A YL A+ +P FA AWSNLA +M G L A Q
Sbjct: 186 PLLVDAHSNLGNLMKAQGLIHEAYSCYLEAVRIQPTFAIAWSNLAGLFMESGDLNRALQY 245
Query: 741 CRQALAINPLMVDAHSNLGNLMKAQGLVQEA 833
++A+ + P DA+ NLGN+ KA G EA
Sbjct: 246 YKEAVKLKPAFPDAYLNLGNVYKALGRPTEA 276
Score = 76.3 bits (186), Expect = 5e-13
Identities = 41/116 (35%), Positives = 68/116 (58%)
Frame = +3
Query: 486 LGAIYYQLHDFDMCVAKNEEALRVEPHFAECYGNMANALKEKGNIDLAIRYYLIAIEFRP 665
L ++ + D + + +EA++++P F + Y N+ N K G AI Y A++ RP
Sbjct: 229 LAGLFMESGDLNRALQYYKEAVKLKPAFPDAYLNLGNVYKALGRPTEAIMCYQHALQMRP 288
Query: 666 NFADAWSNLASAYMRKGRLTEAAQCCRQALAINPLMVDAHSNLGNLMKAQGLVQEA 833
N A A+ N+AS Y +G+L A + +QAL+ +P ++A++NLGN +K G V EA
Sbjct: 289 NSAMAFGNIASIYYEQGQLDLAIRHYKQALSRDPRFLEAYNNLGNALKDIGRVDEA 344
Score = 75.9 bits (185), Expect = 7e-13
Identities = 47/153 (30%), Positives = 74/153 (47%), Gaps = 3/153 (1%)
Frame = +3
Query: 369 DLHLSLAHQMYKSGSYKKALEHSNIVYDRNPLRTDNLLLLGAIYYQ---LHDFDMCVAKN 539
D +LA + G +A + NPL D LG + +H+ C
Sbjct: 156 DAWSNLASAYMRKGRLSEATQCCQQALSLNPLLVDAHSNLGNLMKAQGLIHEAYSCYL-- 213
Query: 540 EEALRVEPHFAECYGNMANALKEKGNIDLAIRYYLIAIEFRPNFADAWSNLASAYMRKGR 719
EA+R++P FA + N+A E G+++ A++YY A++ +P F DA+ NL + Y GR
Sbjct: 214 -EAVRIQPTFAIAWSNLAGLFMESGDLNRALQYYKEAVKLKPAFPDAYLNLGNVYKALGR 272
Query: 720 LTEAAQCCRQALAINPLMVDAHSNLGNLMKAQG 818
TEA C + AL + P A N+ ++ QG
Sbjct: 273 PTEAIMCYQHALQMRPNSAMAFGNIASIYYEQG 305
Score = 73.2 bits (178), Expect = 5e-12
Identities = 44/150 (29%), Positives = 66/150 (43%), Gaps = 34/150 (22%)
Frame = +3
Query: 486 LGAIYYQLHDFDMCVAKNEEALRVEPHFAECYGNMANALKEKGNIDLAIRYYLIAIEFRP 665
+ +IYY+ D+ + ++AL +P F E Y N+ NALK+ G +D A+R Y + +P
Sbjct: 297 IASIYYEQGQLDLAIRHYKQALSRDPRFLEAYNNLGNALKDIGRVDEAVRCYNQCLALQP 356
Query: 666 NFADAWSNLASAYM----------------------------------RKGRLTEAAQCC 743
N A +NL + YM ++G ++A C
Sbjct: 357 NHPQAMANLGNIYMEWNMMGPASSLFKATLAVTTGLSAPFNNLAIIYKQQGNYSDAISCY 416
Query: 744 RQALAINPLMVDAHSNLGNLMKAQGLVQEA 833
+ L I+PL DA N GN K G V EA
Sbjct: 417 NEVLRIDPLAADALVNRGNTYKEIGRVTEA 446
Score = 68.2 bits (165), Expect = 1e-10
Identities = 39/103 (37%), Positives = 55/103 (52%)
Frame = +3
Query: 486 LGAIYYQLHDFDMCVAKNEEALRVEPHFAECYGNMANALKEKGNIDLAIRYYLIAIEFRP 665
L IY Q ++ ++ E LR++P A+ N N KE G + AI+ Y+ AI FRP
Sbjct: 399 LAIIYKQQGNYSDAISCYNEVLRIDPLAADALVNRGNTYKEIGRVTEAIQDYMHAINFRP 458
Query: 666 NFADAWSNLASAYMRKGRLTEAAQCCRQALAINPLMVDAHSNL 794
A+A +NLASAY G + A +QAL + P +A NL
Sbjct: 459 TMAEAHANLASAYKDSGHVEAAITSYKQALLLRPDFPEATCNL 501
Score = 59.3 bits (142), Expect = 7e-08
Identities = 48/180 (26%), Positives = 75/180 (41%)
Frame = +3
Query: 294 RSPLSRNPRSLFCPCAAHDSSEVDEDLHLSLAHQMYKSGSYKKALEHSNIVYDRNPLRTD 473
+ LSR+PR L + + +L + + G +A+ N P
Sbjct: 315 KQALSRDPRFL--------------EAYNNLGNALKDIGRVDEAVRCYNQCLALQPNHPQ 360
Query: 474 NLLLLGAIYYQLHDFDMCVAKNEEALRVEPHFAECYGNMANALKEKGNIDLAIRYYLIAI 653
+ LG IY + + + + L V + + N+A K++GN AI Y +
Sbjct: 361 AMANLGNIYMEWNMMGPASSLFKATLAVTTGLSAPFNNLAIIYKQQGNYSDAISCYNEVL 420
Query: 654 EFRPNFADAWSNLASAYMRKGRLTEAAQCCRQALAINPLMVDAHSNLGNLMKAQGLVQEA 833
P ADA N + Y GR+TEA Q A+ P M +AH+NL + K G V+ A
Sbjct: 421 RIDPLAADALVNRGNTYKEIGRVTEAIQDYMHAINFRPTMAEAHANLASAYKDSGHVEAA 480
Score = 53.9 bits (128), Expect = 3e-06
Identities = 30/97 (30%), Positives = 49/97 (49%)
Frame = +3
Query: 402 KSGSYKKALEHSNIVYDRNPLRTDNLLLLGAIYYQLHDFDMCVAKNEEALRVEPHFAECY 581
+ G+Y A+ N V +PL D L+ G Y ++ + A+ P AE +
Sbjct: 405 QQGNYSDAISCYNEVLRIDPLAADALVNRGNTYKEIGRVTEAIQDYMHAINFRPTMAEAH 464
Query: 582 GNMANALKEKGNIDLAIRYYLIAIEFRPNFADAWSNL 692
N+A+A K+ G+++ AI Y A+ RP+F +A NL
Sbjct: 465 ANLASAYKDSGHVEAAITSYKQALLLRPDFPEATCNL 501
>dbj|BAB92672.1| putative O-linked GlcNAc transferase [Oryza sativa (japonica
cultivar-group)]
Length = 913
Score = 208 bits (530), Expect = 7e-53
Identities = 94/162 (58%), Positives = 128/162 (78%)
Frame = +3
Query: 348 DSSEVDEDLHLSLAHQMYKSGSYKKALEHSNIVYDRNPLRTDNLLLLGAIYYQLHDFDMC 527
D EVDE L +A Q Y++G YK ALEH N VY NP +NLLLLGA+YYQL +FDMC
Sbjct: 46 DPLEVDEGARLEIARQSYRAGDYKAALEHCNAVYRANPRLLENLLLLGAVYYQLREFDMC 105
Query: 528 VAKNEEALRVEPHFAECYGNMANALKEKGNIDLAIRYYLIAIEFRPNFADAWSNLASAYM 707
+AKNEEA+ ++P+ EC+ ++ANA +EKG++D AI++Y+ A++ RP FADAW+NLA+AY
Sbjct: 106 IAKNEEAVAIQPNCPECFNSIANAWREKGDVDNAIQFYVHAVQLRPTFADAWTNLANAYT 165
Query: 708 RKGRLTEAAQCCRQALAINPLMVDAHSNLGNLMKAQGLVQEA 833
RKG L++AA+CC QALA+NP + DA+ NLG+++KAQGL +EA
Sbjct: 166 RKGNLSQAAECCHQALALNPHLADAYCNLGDVLKAQGLYREA 207
Score = 85.5 bits (210), Expect = 9e-16
Identities = 48/151 (31%), Positives = 74/151 (48%)
Frame = +3
Query: 381 SLAHQMYKSGSYKKALEHSNIVYDRNPLRTDNLLLLGAIYYQLHDFDMCVAKNEEALRVE 560
S+A+ + G A++ P D L Y + + +AL +
Sbjct: 125 SIANAWREKGDVDNAIQFYVHAVQLRPTFADAWTNLANAYTRKGNLSQAAECCHQALALN 184
Query: 561 PHFAECYGNMANALKEKGNIDLAIRYYLIAIEFRPNFADAWSNLASAYMRKGRLTEAAQC 740
PH A+ Y N+ + LK +G A +YL A+ +P FA+AW+N+A M+ G +AA
Sbjct: 185 PHLADAYCNLGDVLKAQGLYREAYSHYLDALNIKPTFANAWNNIAGLLMQWGDFNKAALY 244
Query: 741 CRQALAINPLMVDAHSNLGNLMKAQGLVQEA 833
++A+ NP DAH NLGNL K G+ Q+A
Sbjct: 245 YKEAIKCNPAFYDAHLNLGNLYKVTGMRQDA 275
Score = 68.2 bits (165), Expect = 1e-10
Identities = 47/154 (30%), Positives = 73/154 (46%), Gaps = 3/154 (1%)
Frame = +3
Query: 381 SLAHQMYKSGSYKKALEHSNIVYDRNPLRTDNLLLLGAIYYQLHDFDMCVAKNEEALRVE 560
++A + + G + KA + NP D L LG +Y + + A R +
Sbjct: 227 NIAGLLMQWGDFNKAALYYKEAIKCNPAFYDAHLNLGNLYKVTGMRQDAIVCFQNAARAK 286
Query: 561 PHFAECY---GNMANALKEKGNIDLAIRYYLIAIEFRPNFADAWSNLASAYMRKGRLTEA 731
P A Y GN+ NA E+G +DLAI Y AI ++ +A++NL +A GR EA
Sbjct: 287 PENAVAYVFSGNLGNAYHEQGQLDLAILSYRQAIHCNSSYVEAYNNLGNALKDAGRNEEA 346
Query: 732 AQCCRQALAINPLMVDAHSNLGNLMKAQGLVQEA 833
C + LA+ P A +NLGN+ + ++ A
Sbjct: 347 ISCYQTCLALQPSHPQALTNLGNVYMERNMMDIA 380
Score = 62.4 bits (150), Expect = 8e-09
Identities = 38/112 (33%), Positives = 56/112 (49%)
Frame = +3
Query: 486 LGAIYYQLHDFDMCVAKNEEALRVEPHFAECYGNMANALKEKGNIDLAIRYYLIAIEFRP 665
L IY Q + + + E LR++P A+C N N KE G I AI+ Y A+ RP
Sbjct: 401 LAMIYKQQGNCNHAITCFNEVLRIDPMAADCLVNRGNTFKEAGRITEAIQDYFHAVTIRP 460
Query: 666 NFADAWSNLASAYMRKGRLTEAAQCCRQALAINPLMVDAHSNLGNLMKAQGL 821
A+A +NLA+AY G L + +QAL + +A NL + ++ L
Sbjct: 461 TMAEAHANLAAAYKDTGLLEASIISYKQALQLRQDFPEATCNLLHTLQMSSL 512
Score = 62.4 bits (150), Expect = 8e-09
Identities = 40/149 (26%), Positives = 67/149 (44%)
Frame = +3
Query: 381 SLAHQMYKSGSYKKALEHSNIVYDRNPLRTDNLLLLGAIYYQLHDFDMCVAKNEEALRVE 560
+L + + +G ++A+ P L LG +Y + + D+ + L V
Sbjct: 332 NLGNALKDAGRNEEAISCYQTCLALQPSHPQALTNLGNVYMERNMMDIAASLYMATLTVT 391
Query: 561 PHFAECYGNMANALKEKGNIDLAIRYYLIAIEFRPNFADAWSNLASAYMRKGRLTEAAQC 740
+ Y N+A K++GN + AI + + P AD N + + GR+TEA Q
Sbjct: 392 TGLSAPYNNLAMIYKQQGNCNHAITCFNEVLRIDPMAADCLVNRGNTFKEAGRITEAIQD 451
Query: 741 CRQALAINPLMVDAHSNLGNLMKAQGLVQ 827
A+ I P M +AH+NL K GL++
Sbjct: 452 YFHAVTIRPTMAEAHANLAAAYKDTGLLE 480
Score = 59.7 bits (143), Expect = 5e-08
Identities = 30/111 (27%), Positives = 52/111 (46%)
Frame = +3
Query: 486 LGAIYYQLHDFDMCVAKNEEALRVEPHFAECYGNMANALKEKGNIDLAIRYYLIAIEFRP 665
LG Y++ D+ + +A+ + E Y N+ NALK+ G + AI Y + +P
Sbjct: 299 LGNAYHEQGQLDLAILSYRQAIHCNSSYVEAYNNLGNALKDAGRNEEAISCYQTCLALQP 358
Query: 666 NFADAWSNLASAYMRKGRLTEAAQCCRQALAINPLMVDAHSNLGNLMKAQG 818
+ A +NL + YM + + AA L + + ++NL + K QG
Sbjct: 359 SHPQALTNLGNVYMERNMMDIAASLYMATLTVTTGLSAPYNNLAMIYKQQG 409
>pir||E88499 protein K04G7.3 [imported] - Caenorhabditis elegans
Length = 1194
Score = 108 bits (271), Expect = 7e-23
Identities = 57/150 (38%), Positives = 90/150 (60%)
Frame = +3
Query: 384 LAHQMYKSGSYKKALEHSNIVYDRNPLRTDNLLLLGAIYYQLHDFDMCVAKNEEALRVEP 563
LAH+ ++SG+Y +A ++ N+V+ +P LLLL AI +Q + + + + A++V
Sbjct: 174 LAHRQFQSGNYVEAEKYCNLVFQSDPNNLPTLLLLSAINFQTKNLEKSMQYSMLAIKVNN 233
Query: 564 HFAECYGNMANALKEKGNIDLAIRYYLIAIEFRPNFADAWSNLASAYMRKGRLTEAAQCC 743
AE Y N+ N KEKG + A+ Y +A++ +P F DA+ NLA+A + G L +A
Sbjct: 234 QCAEAYSNLGNYYKEKGQLQDALENYKLAVKLKPEFIDAYINLAAALVSGGDLEQAVTAY 293
Query: 744 RQALAINPLMVDAHSNLGNLMKAQGLVQEA 833
AL INP + S+LGNL+KA G ++EA
Sbjct: 294 FNALQINPDLYCVRSDLGNLLKAMGRLEEA 323
Score = 87.0 bits (214), Expect = 3e-16
Identities = 42/116 (36%), Positives = 73/116 (62%)
Frame = +3
Query: 486 LGAIYYQLHDFDMCVAKNEEALRVEPHFAECYGNMANALKEKGNIDLAIRYYLIAIEFRP 665
L +YY+ D+ + ++A+ ++PHF + Y N+ANALKEKG++ A + Y+ A+E P
Sbjct: 412 LACVYYEQGLIDLAIDTYKKAIDLQPHFPDAYCNLANALKEKGSVVEAEQMYMKALELCP 471
Query: 666 NFADAWSNLASAYMRKGRLTEAAQCCRQALAINPLMVDAHSNLGNLMKAQGLVQEA 833
AD+ +NLA+ +G++ +A + +AL I P AHSNL ++++ QG + +A
Sbjct: 472 THADSQNNLANIKREQGKIEDATRLYLKALEIYPEFAAAHSNLASILQQQGKLNDA 527
Score = 83.2 bits (204), Expect = 4e-15
Identities = 48/137 (35%), Positives = 74/137 (53%)
Frame = +3
Query: 423 ALEHSNIVYDRNPLRTDNLLLLGAIYYQLHDFDMCVAKNEEALRVEPHFAECYGNMANAL 602
A+ H +P D + LG + + FD V+ AL + + A +GN+A
Sbjct: 357 AIHHFEKAVTLDPNFLDAYINLGNVLKEARIFDRAVSAYLRALNLSGNHAVVHGNLACVY 416
Query: 603 KEKGNIDLAIRYYLIAIEFRPNFADAWSNLASAYMRKGRLTEAAQCCRQALAINPLMVDA 782
E+G IDLAI Y AI+ +P+F DA+ NLA+A KG + EA Q +AL + P D+
Sbjct: 417 YEQGLIDLAIDTYKKAIDLQPHFPDAYCNLANALKEKGSVVEAEQMYMKALELCPTHADS 476
Query: 783 HSNLGNLMKAQGLVQEA 833
+NL N+ + QG +++A
Sbjct: 477 QNNLANIKREQGKIEDA 493
Score = 80.9 bits (198), Expect = 2e-14
Identities = 48/155 (30%), Positives = 79/155 (50%)
Frame = +3
Query: 369 DLHLSLAHQMYKSGSYKKALEHSNIVYDRNPLRTDNLLLLGAIYYQLHDFDMCVAKNEEA 548
D + +LA+ + + GS +A + + P D+ L I + + +A
Sbjct: 441 DAYCNLANALKEKGSVVEAEQMYMKALELCPTHADSQNNLANIKREQGKIEDATRLYLKA 500
Query: 549 LRVEPHFAECYGNMANALKEKGNIDLAIRYYLIAIEFRPNFADAWSNLASAYMRKGRLTE 728
L + P FA + N+A+ L+++G ++ AI +Y AI P FADA+SN+ + G +
Sbjct: 501 LEIYPEFAAAHSNLASILQQQGKLNDAILHYKEAIRIAPTFADAYSNMGNTLKEMGDSSA 560
Query: 729 AAQCCRQALAINPLMVDAHSNLGNLMKAQGLVQEA 833
A C +A+ INP DAHSNL ++ K G + EA
Sbjct: 561 AIACYNRAIQINPAFADAHSNLASIHKDAGNMAEA 595
Score = 77.0 bits (188), Expect = 3e-13
Identities = 50/129 (38%), Positives = 65/129 (49%)
Frame = +3
Query: 414 YKKALEHSNIVYDRNPLRTDNLLLLGAIYYQLHDFDMCVAKNEEALRVEPHFAECYGNMA 593
Y KALE +Y NL +I Q + + +EA+R+ P FA+ Y NM
Sbjct: 497 YLKALE----IYPEFAAAHSNL---ASILQQQGKLNDAILHYKEAIRIAPTFADAYSNMG 549
Query: 594 NALKEKGNIDLAIRYYLIAIEFRPNFADAWSNLASAYMRKGRLTEAAQCCRQALAINPLM 773
N LKE G+ AI Y AI+ P FADA SNLAS + G + EA Q AL + P
Sbjct: 550 NTLKEMGDSSAAIACYNRAIQINPAFADAHSNLASIHKDAGNMAEAIQSYSTALKLKPDF 609
Query: 774 VDAHSNLGN 800
DA+ NL +
Sbjct: 610 PDAYCNLAH 618
Score = 70.9 bits (172), Expect = 2e-11
Identities = 44/155 (28%), Positives = 73/155 (46%)
Frame = +3
Query: 369 DLHLSLAHQMYKSGSYKKALEHSNIVYDRNPLRTDNLLLLGAIYYQLHDFDMCVAKNEEA 548
+ + +L + + G + ALE+ + P D + L A D + V A
Sbjct: 237 EAYSNLGNYYKEKGQLQDALENYKLAVKLKPEFIDAYINLAAALVSGGDLEQAVTAYFNA 296
Query: 549 LRVEPHFAECYGNMANALKEKGNIDLAIRYYLIAIEFRPNFADAWSNLASAYMRKGRLTE 728
L++ P ++ N LK G ++ A YL AIE +P FA AWSNL + +G +
Sbjct: 297 LQINPDLYCVRSDLGNLLKAMGRLEEAKVCYLKAIETQPQFAVAWSNLGCVFNSQGEIWL 356
Query: 729 AAQCCRQALAINPLMVDAHSNLGNLMKAQGLVQEA 833
A +A+ ++P +DA+ NLGN++K + A
Sbjct: 357 AIHHFEKAVTLDPNFLDAYINLGNVLKEARIFDRA 391
Score = 67.8 bits (164), Expect = 2e-10
Identities = 37/116 (31%), Positives = 60/116 (50%)
Frame = +3
Query: 486 LGAIYYQLHDFDMCVAKNEEALRVEPHFAECYGNMANALKEKGNIDLAIRYYLIAIEFRP 665
LG ++ + + + E+A+ ++P+F + Y N+ N LKE D A+ YL A+
Sbjct: 344 LGCVFNSQGEIWLAIHHFEKAVTLDPNFLDAYINLGNVLKEARIFDRAVSAYLRALNLSG 403
Query: 666 NFADAWSNLASAYMRKGRLTEAAQCCRQALAINPLMVDAHSNLGNLMKAQGLVQEA 833
N A NLA Y +G + A ++A+ + P DA+ NL N +K +G V EA
Sbjct: 404 NHAVVHGNLACVYYEQGLIDLAIDTYKKAIDLQPHFPDAYCNLANALKEKGSVVEA 459
Score = 57.0 bits (136), Expect = 3e-07
Identities = 31/110 (28%), Positives = 55/110 (49%)
Frame = +3
Query: 375 HLSLAHQMYKSGSYKKALEHSNIVYDRNPLRTDNLLLLGAIYYQLHDFDMCVAKNEEALR 554
H +LA + + G A+ H P D +G ++ D +A A++
Sbjct: 511 HSNLASILQQQGKLNDAILHYKEAIRIAPTFADAYSNMGNTLKEMGDSSAAIACYNRAIQ 570
Query: 555 VEPHFAECYGNMANALKEKGNIDLAIRYYLIAIEFRPNFADAWSNLASAY 704
+ P FA+ + N+A+ K+ GN+ AI+ Y A++ +P+F DA+ NLA +
Sbjct: 571 INPAFADAHSNLASIHKDAGNMAEAIQSYSTALKLKPDFPDAYCNLAHCH 620
>ref|NP_498563.1| O-linked N-acetylglucosamine transferase, nucleocytoplasmic, adds
O-linked GlcNAc on transcription factors and nuclear
pore proteins (128.0 kD) [Caenorhabditis elegans]
gi|20198852|gb|AAA62535.2| Hypothetical protein K04G7.3
[Caenorhabditis elegans]
Length = 1151
Score = 108 bits (271), Expect = 7e-23
Identities = 57/150 (38%), Positives = 90/150 (60%)
Frame = +3
Query: 384 LAHQMYKSGSYKKALEHSNIVYDRNPLRTDNLLLLGAIYYQLHDFDMCVAKNEEALRVEP 563
LAH+ ++SG+Y +A ++ N+V+ +P LLLL AI +Q + + + + A++V
Sbjct: 131 LAHRQFQSGNYVEAEKYCNLVFQSDPNNLPTLLLLSAINFQTKNLEKSMQYSMLAIKVNN 190
Query: 564 HFAECYGNMANALKEKGNIDLAIRYYLIAIEFRPNFADAWSNLASAYMRKGRLTEAAQCC 743
AE Y N+ N KEKG + A+ Y +A++ +P F DA+ NLA+A + G L +A
Sbjct: 191 QCAEAYSNLGNYYKEKGQLQDALENYKLAVKLKPEFIDAYINLAAALVSGGDLEQAVTAY 250
Query: 744 RQALAINPLMVDAHSNLGNLMKAQGLVQEA 833
AL INP + S+LGNL+KA G ++EA
Sbjct: 251 FNALQINPDLYCVRSDLGNLLKAMGRLEEA 280
Score = 87.0 bits (214), Expect = 3e-16
Identities = 42/116 (36%), Positives = 73/116 (62%)
Frame = +3
Query: 486 LGAIYYQLHDFDMCVAKNEEALRVEPHFAECYGNMANALKEKGNIDLAIRYYLIAIEFRP 665
L +YY+ D+ + ++A+ ++PHF + Y N+ANALKEKG++ A + Y+ A+E P
Sbjct: 369 LACVYYEQGLIDLAIDTYKKAIDLQPHFPDAYCNLANALKEKGSVVEAEQMYMKALELCP 428
Query: 666 NFADAWSNLASAYMRKGRLTEAAQCCRQALAINPLMVDAHSNLGNLMKAQGLVQEA 833
AD+ +NLA+ +G++ +A + +AL I P AHSNL ++++ QG + +A
Sbjct: 429 THADSQNNLANIKREQGKIEDATRLYLKALEIYPEFAAAHSNLASILQQQGKLNDA 484
Score = 83.2 bits (204), Expect = 4e-15
Identities = 48/137 (35%), Positives = 74/137 (53%)
Frame = +3
Query: 423 ALEHSNIVYDRNPLRTDNLLLLGAIYYQLHDFDMCVAKNEEALRVEPHFAECYGNMANAL 602
A+ H +P D + LG + + FD V+ AL + + A +GN+A
Sbjct: 314 AIHHFEKAVTLDPNFLDAYINLGNVLKEARIFDRAVSAYLRALNLSGNHAVVHGNLACVY 373
Query: 603 KEKGNIDLAIRYYLIAIEFRPNFADAWSNLASAYMRKGRLTEAAQCCRQALAINPLMVDA 782
E+G IDLAI Y AI+ +P+F DA+ NLA+A KG + EA Q +AL + P D+
Sbjct: 374 YEQGLIDLAIDTYKKAIDLQPHFPDAYCNLANALKEKGSVVEAEQMYMKALELCPTHADS 433
Query: 783 HSNLGNLMKAQGLVQEA 833
+NL N+ + QG +++A
Sbjct: 434 QNNLANIKREQGKIEDA 450
Score = 80.9 bits (198), Expect = 2e-14
Identities = 48/155 (30%), Positives = 79/155 (50%)
Frame = +3
Query: 369 DLHLSLAHQMYKSGSYKKALEHSNIVYDRNPLRTDNLLLLGAIYYQLHDFDMCVAKNEEA 548
D + +LA+ + + GS +A + + P D+ L I + + +A
Sbjct: 398 DAYCNLANALKEKGSVVEAEQMYMKALELCPTHADSQNNLANIKREQGKIEDATRLYLKA 457
Query: 549 LRVEPHFAECYGNMANALKEKGNIDLAIRYYLIAIEFRPNFADAWSNLASAYMRKGRLTE 728
L + P FA + N+A+ L+++G ++ AI +Y AI P FADA+SN+ + G +
Sbjct: 458 LEIYPEFAAAHSNLASILQQQGKLNDAILHYKEAIRIAPTFADAYSNMGNTLKEMGDSSA 517
Query: 729 AAQCCRQALAINPLMVDAHSNLGNLMKAQGLVQEA 833
A C +A+ INP DAHSNL ++ K G + EA
Sbjct: 518 AIACYNRAIQINPAFADAHSNLASIHKDAGNMAEA 552
Score = 77.0 bits (188), Expect = 3e-13
Identities = 50/129 (38%), Positives = 65/129 (49%)
Frame = +3
Query: 414 YKKALEHSNIVYDRNPLRTDNLLLLGAIYYQLHDFDMCVAKNEEALRVEPHFAECYGNMA 593
Y KALE +Y NL +I Q + + +EA+R+ P FA+ Y NM
Sbjct: 454 YLKALE----IYPEFAAAHSNL---ASILQQQGKLNDAILHYKEAIRIAPTFADAYSNMG 506
Query: 594 NALKEKGNIDLAIRYYLIAIEFRPNFADAWSNLASAYMRKGRLTEAAQCCRQALAINPLM 773
N LKE G+ AI Y AI+ P FADA SNLAS + G + EA Q AL + P
Sbjct: 507 NTLKEMGDSSAAIACYNRAIQINPAFADAHSNLASIHKDAGNMAEAIQSYSTALKLKPDF 566
Query: 774 VDAHSNLGN 800
DA+ NL +
Sbjct: 567 PDAYCNLAH 575
Score = 70.9 bits (172), Expect = 2e-11
Identities = 44/155 (28%), Positives = 73/155 (46%)
Frame = +3
Query: 369 DLHLSLAHQMYKSGSYKKALEHSNIVYDRNPLRTDNLLLLGAIYYQLHDFDMCVAKNEEA 548
+ + +L + + G + ALE+ + P D + L A D + V A
Sbjct: 194 EAYSNLGNYYKEKGQLQDALENYKLAVKLKPEFIDAYINLAAALVSGGDLEQAVTAYFNA 253
Query: 549 LRVEPHFAECYGNMANALKEKGNIDLAIRYYLIAIEFRPNFADAWSNLASAYMRKGRLTE 728
L++ P ++ N LK G ++ A YL AIE +P FA AWSNL + +G +
Sbjct: 254 LQINPDLYCVRSDLGNLLKAMGRLEEAKVCYLKAIETQPQFAVAWSNLGCVFNSQGEIWL 313
Query: 729 AAQCCRQALAINPLMVDAHSNLGNLMKAQGLVQEA 833
A +A+ ++P +DA+ NLGN++K + A
Sbjct: 314 AIHHFEKAVTLDPNFLDAYINLGNVLKEARIFDRA 348
Score = 67.8 bits (164), Expect = 2e-10
Identities = 37/116 (31%), Positives = 60/116 (50%)
Frame = +3
Query: 486 LGAIYYQLHDFDMCVAKNEEALRVEPHFAECYGNMANALKEKGNIDLAIRYYLIAIEFRP 665
LG ++ + + + E+A+ ++P+F + Y N+ N LKE D A+ YL A+
Sbjct: 301 LGCVFNSQGEIWLAIHHFEKAVTLDPNFLDAYINLGNVLKEARIFDRAVSAYLRALNLSG 360
Query: 666 NFADAWSNLASAYMRKGRLTEAAQCCRQALAINPLMVDAHSNLGNLMKAQGLVQEA 833
N A NLA Y +G + A ++A+ + P DA+ NL N +K +G V EA
Sbjct: 361 NHAVVHGNLACVYYEQGLIDLAIDTYKKAIDLQPHFPDAYCNLANALKEKGSVVEA 416
Score = 57.0 bits (136), Expect = 3e-07
Identities = 31/110 (28%), Positives = 55/110 (49%)
Frame = +3
Query: 375 HLSLAHQMYKSGSYKKALEHSNIVYDRNPLRTDNLLLLGAIYYQLHDFDMCVAKNEEALR 554
H +LA + + G A+ H P D +G ++ D +A A++
Sbjct: 468 HSNLASILQQQGKLNDAILHYKEAIRIAPTFADAYSNMGNTLKEMGDSSAAIACYNRAIQ 527
Query: 555 VEPHFAECYGNMANALKEKGNIDLAIRYYLIAIEFRPNFADAWSNLASAY 704
+ P FA+ + N+A+ K+ GN+ AI+ Y A++ +P+F DA+ NLA +
Sbjct: 528 INPAFADAHSNLASIHKDAGNMAEAIQSYSTALKLKPDFPDAYCNLAHCH 577
>gb|AAB63465.1| O-linked GlcNAc transferase [Caenorhabditis elegans]
Length = 1151
Score = 108 bits (271), Expect = 7e-23
Identities = 57/150 (38%), Positives = 90/150 (60%)
Frame = +3
Query: 384 LAHQMYKSGSYKKALEHSNIVYDRNPLRTDNLLLLGAIYYQLHDFDMCVAKNEEALRVEP 563
LAH+ ++SG+Y +A ++ N+V+ +P LLLL AI +Q + + + + A++V
Sbjct: 131 LAHRQFQSGNYVEAEKYCNLVFQSDPNNLPTLLLLSAINFQTKNLEKSMQYSMLAIKVNN 190
Query: 564 HFAECYGNMANALKEKGNIDLAIRYYLIAIEFRPNFADAWSNLASAYMRKGRLTEAAQCC 743
AE Y N+ N KEKG + A+ Y +A++ +P F DA+ NLA+A + G L +A
Sbjct: 191 QCAEAYSNLGNYYKEKGQLQDALENYKLAVKLKPEFIDAYINLAAALVSGGDLEQAVTAY 250
Query: 744 RQALAINPLMVDAHSNLGNLMKAQGLVQEA 833
AL INP + S+LGNL+KA G ++EA
Sbjct: 251 FNALQINPDLYCVRSDLGNLLKAMGRLEEA 280
Score = 87.0 bits (214), Expect = 3e-16
Identities = 42/116 (36%), Positives = 73/116 (62%)
Frame = +3
Query: 486 LGAIYYQLHDFDMCVAKNEEALRVEPHFAECYGNMANALKEKGNIDLAIRYYLIAIEFRP 665
L +YY+ D+ + ++A+ ++PHF + Y N+ANALKEKG++ A + Y+ A+E P
Sbjct: 369 LACVYYEQGLIDLAIDTYKKAIDLQPHFPDAYCNLANALKEKGSVVEAEQMYMKALELCP 428
Query: 666 NFADAWSNLASAYMRKGRLTEAAQCCRQALAINPLMVDAHSNLGNLMKAQGLVQEA 833
AD+ +NLA+ +G++ +A + +AL I P AHSNL ++++ QG + +A
Sbjct: 429 THADSQNNLANIKREQGKIEDATRLYLKALEIYPEFAAAHSNLASILQQQGKLNDA 484
Score = 83.2 bits (204), Expect = 4e-15
Identities = 48/137 (35%), Positives = 74/137 (53%)
Frame = +3
Query: 423 ALEHSNIVYDRNPLRTDNLLLLGAIYYQLHDFDMCVAKNEEALRVEPHFAECYGNMANAL 602
A+ H +P D + LG + + FD V+ AL + + A +GN+A
Sbjct: 314 AIHHFEKAVTLDPNFLDAYINLGNVLKEARIFDRAVSAYLRALNLSGNHAVVHGNLACVY 373
Query: 603 KEKGNIDLAIRYYLIAIEFRPNFADAWSNLASAYMRKGRLTEAAQCCRQALAINPLMVDA 782
E+G IDLAI Y AI+ +P+F DA+ NLA+A KG + EA Q +AL + P D+
Sbjct: 374 YEQGLIDLAIDTYKKAIDLQPHFPDAYCNLANALKEKGSVVEAEQMYMKALELCPTHADS 433
Query: 783 HSNLGNLMKAQGLVQEA 833
+NL N+ + QG +++A
Sbjct: 434 QNNLANIKREQGKIEDA 450
Score = 80.9 bits (198), Expect = 2e-14
Identities = 48/155 (30%), Positives = 79/155 (50%)
Frame = +3
Query: 369 DLHLSLAHQMYKSGSYKKALEHSNIVYDRNPLRTDNLLLLGAIYYQLHDFDMCVAKNEEA 548
D + +LA+ + + GS +A + + P D+ L I + + +A
Sbjct: 398 DAYCNLANALKEKGSVVEAEQMYMKALELCPTHADSQNNLANIKREQGKIEDATRLYLKA 457
Query: 549 LRVEPHFAECYGNMANALKEKGNIDLAIRYYLIAIEFRPNFADAWSNLASAYMRKGRLTE 728
L + P FA + N+A+ L+++G ++ AI +Y AI P FADA+SN+ + G +
Sbjct: 458 LEIYPEFAAAHSNLASILQQQGKLNDAILHYKEAIRIAPTFADAYSNMGNTLKEMGDSSA 517
Query: 729 AAQCCRQALAINPLMVDAHSNLGNLMKAQGLVQEA 833
A C +A+ INP DAHSNL ++ K G + EA
Sbjct: 518 AIACYNRAIQINPAFADAHSNLASIHKDAGNMAEA 552
Score = 77.0 bits (188), Expect = 3e-13
Identities = 50/129 (38%), Positives = 65/129 (49%)
Frame = +3
Query: 414 YKKALEHSNIVYDRNPLRTDNLLLLGAIYYQLHDFDMCVAKNEEALRVEPHFAECYGNMA 593
Y KALE +Y NL +I Q + + +EA+R+ P FA+ Y NM
Sbjct: 454 YLKALE----IYPEFAAAHSNL---ASILQQQGKLNDAILHYKEAIRIAPTFADAYSNMG 506
Query: 594 NALKEKGNIDLAIRYYLIAIEFRPNFADAWSNLASAYMRKGRLTEAAQCCRQALAINPLM 773
N LKE G+ AI Y AI+ P FADA SNLAS + G + EA Q AL + P
Sbjct: 507 NTLKEMGDSSAAIACYNRAIQINPAFADAHSNLASIHKDAGNMAEAIQSYSTALKLKPDF 566
Query: 774 VDAHSNLGN 800
DA+ NL +
Sbjct: 567 PDAYCNLAH 575
Score = 70.9 bits (172), Expect = 2e-11
Identities = 44/155 (28%), Positives = 73/155 (46%)
Frame = +3
Query: 369 DLHLSLAHQMYKSGSYKKALEHSNIVYDRNPLRTDNLLLLGAIYYQLHDFDMCVAKNEEA 548
+ + +L + + G + ALE+ + P D + L A D + V A
Sbjct: 194 EAYSNLGNYYKEKGQLQDALENYKLAVKLKPEFIDAYINLAAALVSGGDLEQAVTAYFNA 253
Query: 549 LRVEPHFAECYGNMANALKEKGNIDLAIRYYLIAIEFRPNFADAWSNLASAYMRKGRLTE 728
L++ P ++ N LK G ++ A YL AIE +P FA AWSNL + +G +
Sbjct: 254 LQINPDLYCVRSDLGNLLKAMGRLEEAKVCYLKAIETQPQFAVAWSNLGCVFNSQGEIWL 313
Query: 729 AAQCCRQALAINPLMVDAHSNLGNLMKAQGLVQEA 833
A +A+ ++P +DA+ NLGN++K + A
Sbjct: 314 AIHHFEKAVTLDPNFLDAYINLGNVLKEARIFDRA 348
Score = 67.8 bits (164), Expect = 2e-10
Identities = 37/116 (31%), Positives = 60/116 (50%)
Frame = +3
Query: 486 LGAIYYQLHDFDMCVAKNEEALRVEPHFAECYGNMANALKEKGNIDLAIRYYLIAIEFRP 665
LG ++ + + + E+A+ ++P+F + Y N+ N LKE D A+ YL A+
Sbjct: 301 LGCVFNSQGEIWLAIHHFEKAVTLDPNFLDAYINLGNVLKEARIFDRAVSAYLRALNLSG 360
Query: 666 NFADAWSNLASAYMRKGRLTEAAQCCRQALAINPLMVDAHSNLGNLMKAQGLVQEA 833
N A NLA Y +G + A ++A+ + P DA+ NL N +K +G V EA
Sbjct: 361 NHAVVHGNLACVYYEQGLIDLAIDTYKKAIDLQPHFPDAYCNLANALKEKGSVVEA 416
Score = 57.0 bits (136), Expect = 3e-07
Identities = 31/110 (28%), Positives = 55/110 (49%)
Frame = +3
Query: 375 HLSLAHQMYKSGSYKKALEHSNIVYDRNPLRTDNLLLLGAIYYQLHDFDMCVAKNEEALR 554
H +LA + + G A+ H P D +G ++ D +A A++
Sbjct: 468 HSNLASILQQQGKLNDAILHYKEAIRIAPTFADAYSNMGNTLKEMGDSSAAIACYNRAIQ 527
Query: 555 VEPHFAECYGNMANALKEKGNIDLAIRYYLIAIEFRPNFADAWSNLASAY 704
+ P FA+ + N+A+ K+ GN+ AI+ Y A++ +P+F DA+ NLA +
Sbjct: 528 INPAFADAHSNLASIHKDAGNMAEAIQSYSTALKLKPDFPDAYCNLAHCH 577
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 738,014,759
Number of Sequences: 1393205
Number of extensions: 17383731
Number of successful extensions: 141778
Number of sequences better than 10.0: 1080
Number of HSP's better than 10.0 without gapping: 77497
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 127359
length of database: 448,689,247
effective HSP length: 122
effective length of database: 278,718,237
effective search space used: 43201326735
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)