Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
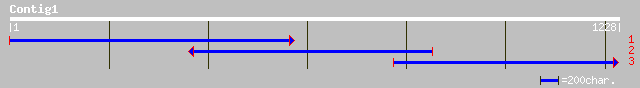
Query= KMC006212A_C01 KMC006212A_c01
(1227 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_565958.1| putative villin 2; protein id: At2g41740.1, sup... 444 e-123
pir||E84845 probable villin 2 [imported] - Arabidopsis thaliana 444 e-123
gb|AAM13051.1| unknown protein [Arabidopsis thaliana] gi|2213650... 443 e-123
pir||T46177 villin 3 homolog T8H10.10 - Arabidopsis thaliana (fr... 443 e-123
sp|O81645|VIL3_ARATH Villin 3 gi|11358922|pir||T50668 villin 3 [... 443 e-123
>ref|NP_565958.1| putative villin 2; protein id: At2g41740.1, supported by cDNA:
gi_19310557, supported by cDNA: gi_3415114 [Arabidopsis
thaliana] gi|25091521|sp|O81644|VIL2_ARATH Villin 2
gi|19310558|gb|AAL85012.1| putative villin 2 protein
[Arabidopsis thaliana] gi|20196894|gb|AAC02774.2|
putative villin 2 [Arabidopsis thaliana]
gi|22136974|gb|AAM91716.1| putative villin 2 protein
[Arabidopsis thaliana]
Length = 976
Score = 444 bits (1142), Expect = e-123
Identities = 228/307 (74%), Positives = 248/307 (80%), Gaps = 2/307 (0%)
Frame = +3
Query: 312 AAKVLDPAFQGVGQRVGTEIWRIENFQPVPLPKSEHGKFYMGDSYIILQTTQGKGGSYLF 491
+ KVLDPAFQG GQ+ GTEIWRIENF+ VP+PKSEHGKFYMGD+YI+LQTTQ KGG+YLF
Sbjct: 2 STKVLDPAFQGAGQKPGTEIWRIENFEAVPVPKSEHGKFYMGDTYIVLQTTQNKGGAYLF 61
Query: 492 DIHFWIGKDTSQDEAGTAAIKTVELDASLGGRAVQHREIQGHESDKFLSYFKPCIIPLEG 671
DIHFWIGKDTSQDEAGTAA+KTVELDA LGGRAVQHREIQGHESDKFLSYFKPCIIPLEG
Sbjct: 62 DIHFWIGKDTSQDEAGTAAVKTVELDAVLGGRAVQHREIQGHESDKFLSYFKPCIIPLEG 121
Query: 672 GVASGFKTPEEEEFETRLYVCKGKRVVRIKQVPFARSSLNHDDVFILDTQDKIYQFNGAN 851
GVASGFKT EEE FETRLY CKGKR +R+KQVPFARSSLNHDDVFILDT++KIYQFNGAN
Sbjct: 122 GVASGFKTVEEEVFETRLYTCKGKRAIRLKQVPFARSSLNHDDVFILDTEEKIYQFNGAN 181
Query: 852 SNIQERAKALEVIQLLKEKYHEGKCDVAIVDDGKLDTESDSGEFWVLFGWFCPHG*KGDR 1031
SNIQERAKALEV+Q LK+KYHEG CDVAIVDDGKLDTESDSG FWVLFG F P G K
Sbjct: 182 SNIQERAKALEVVQYLKDKYHEGTCDVAIVDDGKLDTESDSGAFWVLFGGFAPIGRKVAN 241
Query: 1032 GDDICSRKHFCSNL*LLLMVMSSLVE--GGTFLSPLLENSKLLFTWTVVLRYMSGVGGVT 1205
DDI S L + +E G +LEN+K Y+ VG VT
Sbjct: 242 DDDIVPE----STPPKLYCITDGKMEPIDGDLSKSMLENTKCYLLDCGAEIYI-WVGRVT 296
Query: 1206 QVEERKA 1226
QV+ERKA
Sbjct: 297 QVDERKA 303
Score = 84.7 bits (208), Expect = 3e-15
Identities = 54/190 (28%), Positives = 93/190 (48%), Gaps = 11/190 (5%)
Frame = +3
Query: 366 EIWRIENFQPVPLPKSEHGKFYMGDSYIILQTTQGKGGSYLFDIHFWIGKDTSQDEAGTA 545
E+W + PLPK + GK Y GD Y++L T + + W GK + ++ TA
Sbjct: 393 EVWYVNGKVKTPLPKEDIGKLYSGDCYLVLYTYHSGERKDEYFLSCWFGKKSIPEDQDTA 452
Query: 546 AIKTVELDASLGGRAVQHREIQGHESDKFLSYFKPCIIPLEGGVASGFKTP-EEEEFETR 722
+ SL GR VQ R +G E +F++ F+P ++ L+GG++SG+K+ E E
Sbjct: 453 IRLANTMSNSLKGRPVQGRIYEGKEPPQFVALFQPMVV-LKGGLSSGYKSSMGESESTDE 511
Query: 723 LYVCKGKRVVRIK----------QVPFARSSLNHDDVFILDTQDKIYQFNGANSNIQERA 872
Y + +V++ QV +SLN + F+L + ++ ++G S ++
Sbjct: 512 TYTPESIALVQVSGTGVHNNKAVQVETVATSLNSYECFLLQSGTSMFLWHGNQSTHEQLE 571
Query: 873 KALEVIQLLK 902
A +V + LK
Sbjct: 572 LATKVAEFLK 581
>pir||E84845 probable villin 2 [imported] - Arabidopsis thaliana
Length = 955
Score = 444 bits (1142), Expect = e-123
Identities = 228/307 (74%), Positives = 248/307 (80%), Gaps = 2/307 (0%)
Frame = +3
Query: 312 AAKVLDPAFQGVGQRVGTEIWRIENFQPVPLPKSEHGKFYMGDSYIILQTTQGKGGSYLF 491
+ KVLDPAFQG GQ+ GTEIWRIENF+ VP+PKSEHGKFYMGD+YI+LQTTQ KGG+YLF
Sbjct: 2 STKVLDPAFQGAGQKPGTEIWRIENFEAVPVPKSEHGKFYMGDTYIVLQTTQNKGGAYLF 61
Query: 492 DIHFWIGKDTSQDEAGTAAIKTVELDASLGGRAVQHREIQGHESDKFLSYFKPCIIPLEG 671
DIHFWIGKDTSQDEAGTAA+KTVELDA LGGRAVQHREIQGHESDKFLSYFKPCIIPLEG
Sbjct: 62 DIHFWIGKDTSQDEAGTAAVKTVELDAVLGGRAVQHREIQGHESDKFLSYFKPCIIPLEG 121
Query: 672 GVASGFKTPEEEEFETRLYVCKGKRVVRIKQVPFARSSLNHDDVFILDTQDKIYQFNGAN 851
GVASGFKT EEE FETRLY CKGKR +R+KQVPFARSSLNHDDVFILDT++KIYQFNGAN
Sbjct: 122 GVASGFKTVEEEVFETRLYTCKGKRAIRLKQVPFARSSLNHDDVFILDTEEKIYQFNGAN 181
Query: 852 SNIQERAKALEVIQLLKEKYHEGKCDVAIVDDGKLDTESDSGEFWVLFGWFCPHG*KGDR 1031
SNIQERAKALEV+Q LK+KYHEG CDVAIVDDGKLDTESDSG FWVLFG F P G K
Sbjct: 182 SNIQERAKALEVVQYLKDKYHEGTCDVAIVDDGKLDTESDSGAFWVLFGGFAPIGRKVAN 241
Query: 1032 GDDICSRKHFCSNL*LLLMVMSSLVE--GGTFLSPLLENSKLLFTWTVVLRYMSGVGGVT 1205
DDI S L + +E G +LEN+K Y+ VG VT
Sbjct: 242 DDDIVPE----STPPKLYCITDGKMEPIDGDLSKSMLENTKCYLLDCGAEIYI-WVGRVT 296
Query: 1206 QVEERKA 1226
QV+ERKA
Sbjct: 297 QVDERKA 303
Score = 73.6 bits (179), Expect = 6e-12
Identities = 52/181 (28%), Positives = 83/181 (45%), Gaps = 2/181 (1%)
Frame = +3
Query: 366 EIWRIENFQPVPLPKSEHGKFYMGDSYIILQTTQGKGGSYLFDIHFWIGKDTSQDEAGTA 545
E+W + PLPK + GK Y GD Y++L T + + W GK + ++ TA
Sbjct: 393 EVWYVNGKVKTPLPKEDIGKLYSGDCYLVLYTYHSGERKDEYFLSCWFGKKSIPEDQDTA 452
Query: 546 AIKTVELDASLGGRAVQHREIQGHESDKFLSYFKPCIIPLEGGVASGFKTPEEEEFETRL 725
+ SL GR VQ R +G E +F++ F+P ++ L+ TPE L
Sbjct: 453 IRLANTMSNSLKGRPVQGRIYEGKEPPQFVALFQPMVV-LKSESTDETYTPE----SIAL 507
Query: 726 YVCKGKRVVRIK--QVPFARSSLNHDDVFILDTQDKIYQFNGANSNIQERAKALEVIQLL 899
G V K QV +SLN + F+L + ++ ++G S ++ A +V + L
Sbjct: 508 VQVSGTGVHNNKAVQVETVATSLNSYECFLLQSGTSMFLWHGNQSTHEQLELATKVAEFL 567
Query: 900 K 902
K
Sbjct: 568 K 568
>gb|AAM13051.1| unknown protein [Arabidopsis thaliana] gi|22136508|gb|AAM91332.1|
unknown protein [Arabidopsis thaliana]
Length = 618
Score = 443 bits (1140), Expect = e-123
Identities = 230/317 (72%), Positives = 251/317 (78%), Gaps = 10/317 (3%)
Frame = +3
Query: 303 MSNAAKVLDPAFQGVGQRVGTEIWRIENFQPVPLPKSEHGKFYMGDSYIILQTTQGKGGS 482
MS + KVLDPAFQGVGQ+ GTEIWRIENF+PVP+PKSEHGKFYMGD+YI+LQTTQ KGG+
Sbjct: 1 MSGSTKVLDPAFQGVGQKPGTEIWRIENFEPVPVPKSEHGKFYMGDTYIVLQTTQNKGGA 60
Query: 483 YLFDIHFWIGKDTSQDEAGTAAIKTVELDASLGGRAVQHREIQGHESDKFLSYFKPCIIP 662
YLFDIHFWIGKDTSQDEAGTAA+KTVELDA+LGGRAVQ+REIQGHESDKFLSYFKPCIIP
Sbjct: 61 YLFDIHFWIGKDTSQDEAGTAAVKTVELDAALGGRAVQYREIQGHESDKFLSYFKPCIIP 120
Query: 663 LEGGVASGFKTPEEEEFETRLYVCKGKRVVRIKQVPFARSSLNHDDVFILDTQDKIYQFN 842
LEGGVASGFK PEEEEFETRLY CKGKR V +KQVPFARSSLNHDDVFILDT++KIYQFN
Sbjct: 121 LEGGVASGFKKPEEEEFETRLYTCKGKRAVHLKQVPFARSSLNHDDVFILDTKEKIYQFN 180
Query: 843 GANSNIQERAKALEVIQLLKEKYHEGKCDVAIVDDGKLDTESDSGEFWVLFGWFCPHG*K 1022
GANSNIQERAKAL VIQ LK+K+HEG DVAIVDDGKLDTESDSGEFWVLFG F P K
Sbjct: 181 GANSNIQERAKALVVIQYLKDKFHEGTSDVAIVDDGKLDTESDSGEFWVLFGGFAPIARK 240
Query: 1023 GDRGDDICSRKHFCSNL*LLLMVMSSLVEG--GTFLSPLLENSKL--------LFTWTVV 1172
D+I + L + VE G +LEN+K +F W
Sbjct: 241 VASEDEIIPE----TTPPKLYSIADGQVESIDGDLSKSMLENNKCYLLDCGSEIFIW--- 293
Query: 1173 LRYMSGVGGVTQVEERK 1223
VG VTQVEERK
Sbjct: 294 ------VGRVTQVEERK 304
Score = 80.5 bits (197), Expect = 5e-14
Identities = 55/207 (26%), Positives = 101/207 (48%), Gaps = 11/207 (5%)
Frame = +3
Query: 324 LDPAFQGVGQRVGTEIWRIENFQPVPLPKSEHGKFYMGDSYIILQTTQGKGGSYLFDIHF 503
+ P +G G+ E+W I+ L K GK Y GD Y++L T + +
Sbjct: 384 IPPLLEGGGK---LEVWYIDANSKTVLSKDHVGKLYSGDCYLVLYTYHSGERKEDYFLCC 440
Query: 504 WIGKDTSQDEAGTAAIKTVELDASLGGRAVQHREIQGHESDKFLSYFKPCIIPLEGGVAS 683
W GK+++Q++ TA + SL GR VQ R +G E +F++ F+ ++ L+GG++S
Sbjct: 441 WFGKNSNQEDQETAVRLASTMTNSLKGRPVQARIFEGKEPPQFVALFQHMVV-LKGGLSS 499
Query: 684 GFKTPEEEEFET-RLYVCKGKRVVRIK----------QVPFARSSLNHDDVFILDTQDKI 830
G+K E+ + Y + ++++ QV +SLN D F+L + +
Sbjct: 500 GYKNSMTEKGSSGETYTPESIALIQVSGTGVHNNKALQVEAVATSLNSYDCFLLQSGTSM 559
Query: 831 YQFNGANSNIQERAKALEVIQLLKEKY 911
+ + G +S +++ A +V + LK +
Sbjct: 560 FLWVGNHSTHEQQELAAKVAEFLKSAW 586
>pir||T46177 villin 3 homolog T8H10.10 - Arabidopsis thaliana (fragment)
gi|6706412|emb|CAB66098.1| villin 3 fragment [Arabidopsis
thaliana]
Length = 583
Score = 443 bits (1140), Expect = e-123
Identities = 230/317 (72%), Positives = 251/317 (78%), Gaps = 10/317 (3%)
Frame = +3
Query: 303 MSNAAKVLDPAFQGVGQRVGTEIWRIENFQPVPLPKSEHGKFYMGDSYIILQTTQGKGGS 482
MS + KVLDPAFQGVGQ+ GTEIWRIENF+PVP+PKSEHGKFYMGD+YI+LQTTQ KGG+
Sbjct: 1 MSGSTKVLDPAFQGVGQKPGTEIWRIENFEPVPVPKSEHGKFYMGDTYIVLQTTQNKGGA 60
Query: 483 YLFDIHFWIGKDTSQDEAGTAAIKTVELDASLGGRAVQHREIQGHESDKFLSYFKPCIIP 662
YLFDIHFWIGKDTSQDEAGTAA+KTVELDA+LGGRAVQ+REIQGHESDKFLSYFKPCIIP
Sbjct: 61 YLFDIHFWIGKDTSQDEAGTAAVKTVELDAALGGRAVQYREIQGHESDKFLSYFKPCIIP 120
Query: 663 LEGGVASGFKTPEEEEFETRLYVCKGKRVVRIKQVPFARSSLNHDDVFILDTQDKIYQFN 842
LEGGVASGFK PEEEEFETRLY CKGKR V +KQVPFARSSLNHDDVFILDT++KIYQFN
Sbjct: 121 LEGGVASGFKKPEEEEFETRLYTCKGKRAVHLKQVPFARSSLNHDDVFILDTKEKIYQFN 180
Query: 843 GANSNIQERAKALEVIQLLKEKYHEGKCDVAIVDDGKLDTESDSGEFWVLFGWFCPHG*K 1022
GANSNIQERAKAL VIQ LK+K+HEG DVAIVDDGKLDTESDSGEFWVLFG F P K
Sbjct: 181 GANSNIQERAKALVVIQYLKDKFHEGTSDVAIVDDGKLDTESDSGEFWVLFGGFAPIARK 240
Query: 1023 GDRGDDICSRKHFCSNL*LLLMVMSSLVEG--GTFLSPLLENSKL--------LFTWTVV 1172
D+I + L + VE G +LEN+K +F W
Sbjct: 241 VASEDEIIPE----TTPPKLYSIADGQVESIDGDLSKSMLENNKCYLLDCGSEIFIW--- 293
Query: 1173 LRYMSGVGGVTQVEERK 1223
VG VTQVEERK
Sbjct: 294 ------VGRVTQVEERK 304
Score = 80.1 bits (196), Expect = 7e-14
Identities = 55/204 (26%), Positives = 100/204 (48%), Gaps = 11/204 (5%)
Frame = +3
Query: 324 LDPAFQGVGQRVGTEIWRIENFQPVPLPKSEHGKFYMGDSYIILQTTQGKGGSYLFDIHF 503
+ P +G G+ E+W I+ L K GK Y GD Y++L T + +
Sbjct: 384 IPPLLEGGGK---LEVWYIDANSKTVLSKDHVGKLYSGDCYLVLYTYHSGERKEDYFLCC 440
Query: 504 WIGKDTSQDEAGTAAIKTVELDASLGGRAVQHREIQGHESDKFLSYFKPCIIPLEGGVAS 683
W GK+++Q++ TA + SL GR VQ R +G E +F++ F+ ++ L+GG++S
Sbjct: 441 WFGKNSNQEDQETAVRLASTMTNSLKGRPVQARIFEGKEPPQFVALFQHMVV-LKGGLSS 499
Query: 684 GFKTPEEEEFET-RLYVCKGKRVVRIK----------QVPFARSSLNHDDVFILDTQDKI 830
G+K E+ + Y + ++++ QV +SLN D F+L + +
Sbjct: 500 GYKNSMTEKGSSGETYTPESIALIQVSGTGVHNNKALQVEAVATSLNSYDCFLLQSGTSM 559
Query: 831 YQFNGANSNIQERAKALEVIQLLK 902
+ + G +S +++ A +V + LK
Sbjct: 560 FLWVGNHSTHEQQELAAKVAEFLK 583
>sp|O81645|VIL3_ARATH Villin 3 gi|11358922|pir||T50668 villin 3 [imported] - Arabidopsis
thaliana gi|3415117|gb|AAC31607.1| villin 3 [Arabidopsis
thaliana]
Length = 966
Score = 443 bits (1140), Expect = e-123
Identities = 230/317 (72%), Positives = 251/317 (78%), Gaps = 10/317 (3%)
Frame = +3
Query: 303 MSNAAKVLDPAFQGVGQRVGTEIWRIENFQPVPLPKSEHGKFYMGDSYIILQTTQGKGGS 482
MS + KVLDPAFQGVGQ+ GTEIWRIENF+PVP+PKSEHGKFYMGD+YI+LQTTQ KGG+
Sbjct: 1 MSGSTKVLDPAFQGVGQKPGTEIWRIENFEPVPVPKSEHGKFYMGDTYIVLQTTQNKGGA 60
Query: 483 YLFDIHFWIGKDTSQDEAGTAAIKTVELDASLGGRAVQHREIQGHESDKFLSYFKPCIIP 662
YLFDIHFWIGKDTSQDEAGTAA+KTVELDA+LGGRAVQ+REIQGHESDKFLSYFKPCIIP
Sbjct: 61 YLFDIHFWIGKDTSQDEAGTAAVKTVELDAALGGRAVQYREIQGHESDKFLSYFKPCIIP 120
Query: 663 LEGGVASGFKTPEEEEFETRLYVCKGKRVVRIKQVPFARSSLNHDDVFILDTQDKIYQFN 842
LEGGVASGFK PEEEEFETRLY CKGKR V +KQVPFARSSLNHDDVFILDT++KIYQFN
Sbjct: 121 LEGGVASGFKKPEEEEFETRLYTCKGKRAVHLKQVPFARSSLNHDDVFILDTKEKIYQFN 180
Query: 843 GANSNIQERAKALEVIQLLKEKYHEGKCDVAIVDDGKLDTESDSGEFWVLFGWFCPHG*K 1022
GANSNIQERAKAL VIQ LK+K+HEG DVAIVDDGKLDTESDSGEFWVLFG F P K
Sbjct: 181 GANSNIQERAKALVVIQYLKDKFHEGTSDVAIVDDGKLDTESDSGEFWVLFGGFAPIARK 240
Query: 1023 GDRGDDICSRKHFCSNL*LLLMVMSSLVEG--GTFLSPLLENSKL--------LFTWTVV 1172
D+I + L + VE G +LEN+K +F W
Sbjct: 241 VASEDEIIPE----TTPPKLYSIADGQVESIDGDLSKSMLENNKCYLLDCGSEIFIW--- 293
Query: 1173 LRYMSGVGGVTQVEERK 1223
VG VTQVEERK
Sbjct: 294 ------VGRVTQVEERK 304
Score = 82.0 bits (201), Expect = 2e-14
Identities = 63/236 (26%), Positives = 109/236 (45%), Gaps = 11/236 (4%)
Frame = +3
Query: 324 LDPAFQGVGQRVGTEIWRIENFQPVPLPKSEHGKFYMGDSYIILQTTQGKGGSYLFDIHF 503
+ P +G G+ E+W I+ L K GK Y GD Y++L T + +
Sbjct: 384 IPPLLEGGGK---LEVWYIDANSKTVLSKDHVGKLYSGDCYLVLYTYHSGERKEDYFLCC 440
Query: 504 WIGKDTSQDEAGTAAIKTVELDASLGGRAVQHREIQGHESDKFLSYFKPCIIPLEGGVAS 683
W GK+++Q++ TA + SL GR VQ R +G E +F++ F+ ++ L+GG++S
Sbjct: 441 WFGKNSNQEDQETAVRLASTMTNSLKGRPVQARIFEGKEPPQFVALFQHMVV-LKGGLSS 499
Query: 684 GFKTPEEEEFET-RLYVCKGKRVVRIK----------QVPFARSSLNHDDVFILDTQDKI 830
G+K E+ + Y + ++++ QV +SLN D F+L + +
Sbjct: 500 GYKNSMTEKGSSGETYTPESIALIQVSGTGVHNNKALQVEAVATSLNSYDCFLLQSGTSM 559
Query: 831 YQFNGANSNIQERAKALEVIQLLKEKYHEGKCDVAIVDDGKLDTESDSGEFWVLFG 998
+ + G +S +++ A +V + LK + K TES S FW G
Sbjct: 560 FLWVGNHSTHEQQELAAKVAEFLKPG--------TTIKHAKEGTESSS--FWFALG 605
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,103,570,977
Number of Sequences: 1393205
Number of extensions: 25665441
Number of successful extensions: 94853
Number of sequences better than 10.0: 172
Number of HSP's better than 10.0 without gapping: 80736
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 91228
length of database: 448,689,247
effective HSP length: 126
effective length of database: 273,145,417
effective search space used: 77027007594
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)