Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC005765A_C01 KMC005765A_c01
(488 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
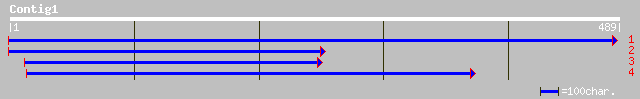
ref|NP_195903.1| putative protein; protein id: At5g02830.1 [Arab... 56 2e-07
dbj|BAC42187.1| unknown protein [Arabidopsis thaliana] 56 2e-07
gb|AAL47991.1| GH23534p [Drosophila melanogaster] 32 5.0
ref|NP_477260.1| Daughters against dpp CG5201-PA gi|24647586|ref... 32 5.0
dbj|BAA22841.1| DAD polypeptide [Drosophila melanogaster] 32 5.0
>ref|NP_195903.1| putative protein; protein id: At5g02830.1 [Arabidopsis thaliana]
gi|11357906|pir||T48304 hypothetical protein F9G14.140 -
Arabidopsis thaliana gi|7413558|emb|CAB86037.1| putative
protein [Arabidopsis thaliana]
Length = 798
Score = 55.8 bits (133), Expect = 2e-07
Identities = 35/87 (40%), Positives = 47/87 (53%)
Frame = -2
Query: 487 KELLEVQGTIIKLLRNELGLEXLPARTRFAPSDTPKFESPDLSNLSIEALPEEKALATTM 308
K+ + VQ ++KLLR+EL L LPA R D + D N K+ +
Sbjct: 721 KQEITVQEALVKLLRDELSLVVLPAGQRNIIQDAHCVDDADQENT--------KSFVSIS 772
Query: 307 GYQTRRPAVLQRLKVTKKSLFSWLQRK 227
TRRPA+L+RL VTK SL+ WLQR+
Sbjct: 773 S--TRRPAILERLMVTKASLYQWLQRR 797
>dbj|BAC42187.1| unknown protein [Arabidopsis thaliana]
Length = 852
Score = 55.8 bits (133), Expect = 2e-07
Identities = 35/87 (40%), Positives = 47/87 (53%)
Frame = -2
Query: 487 KELLEVQGTIIKLLRNELGLEXLPARTRFAPSDTPKFESPDLSNLSIEALPEEKALATTM 308
K+ + VQ ++KLLR+EL L LPA R D + D N K+ +
Sbjct: 775 KQEITVQEALVKLLRDELSLVVLPAGQRNIIQDAHCVDDADQENT--------KSFVSIS 826
Query: 307 GYQTRRPAVLQRLKVTKKSLFSWLQRK 227
TRRPA+L+RL VTK SL+ WLQR+
Sbjct: 827 S--TRRPAILERLMVTKASLYQWLQRR 851
>gb|AAL47991.1| GH23534p [Drosophila melanogaster]
Length = 507
Score = 31.6 bits (70), Expect = 5.0
Identities = 13/25 (52%), Positives = 15/25 (60%)
Frame = -2
Query: 145 DVISPLPAASSTCIFASGYSCISIC 71
DV+ P P+A C A GYSC S C
Sbjct: 54 DVLPPPPSACDRCCTAPGYSCASSC 78
>ref|NP_477260.1| Daughters against dpp CG5201-PA gi|24647586|ref|NP_732196.1|
Daughters against dpp CG5201-PB
gi|7300219|gb|AAF55383.1| CG5201-PB [Drosophila
melanogaster] gi|23171519|gb|AAN13728.1| CG5201-PA
[Drosophila melanogaster]
Length = 568
Score = 31.6 bits (70), Expect = 5.0
Identities = 13/25 (52%), Positives = 15/25 (60%)
Frame = -2
Query: 145 DVISPLPAASSTCIFASGYSCISIC 71
DV+ P P+A C A GYSC S C
Sbjct: 115 DVLPPPPSACDRCCTAPGYSCASSC 139
>dbj|BAA22841.1| DAD polypeptide [Drosophila melanogaster]
Length = 568
Score = 31.6 bits (70), Expect = 5.0
Identities = 13/25 (52%), Positives = 15/25 (60%)
Frame = -2
Query: 145 DVISPLPAASSTCIFASGYSCISIC 71
DV+ P P+A C A GYSC S C
Sbjct: 115 DVLPPPPSACDRCCTAPGYSCASSC 139
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 405,224,967
Number of Sequences: 1393205
Number of extensions: 8409273
Number of successful extensions: 18995
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 18481
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 18986
length of database: 448,689,247
effective HSP length: 113
effective length of database: 291,257,082
effective search space used: 14271597018
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)