Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC005752A_C01 KMC005752A_c01
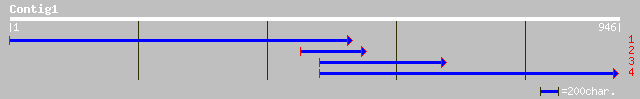
(907 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|XP_141502.1| similar to Formin 1 isoforms I/II/III (Limb def... 37 0.53
gb|AAK16728.1|AF332962_1 ferrochelatase [Chlamydomonas reinhardtii] 36 0.91
dbj|BAB63481.1| P0002B05.1 [Oryza sativa (japonica cultivar-grou... 35 1.6
ref|NP_253244.1| type 4 fimbrial biogenesis protein PilY1 [Pseud... 34 2.7
pir||E71615 probable amine transporter PFB0435c - malaria parasi... 34 3.5
>ref|XP_141502.1| similar to Formin 1 isoforms I/II/III (Limb deformity protein) [Mus
musculus]
Length = 691
Score = 36.6 bits (83), Expect = 0.53
Identities = 25/84 (29%), Positives = 40/84 (46%)
Frame = -3
Query: 716 HRRPSAELSGQTVDSLGKRTTSLNSLPRKDEGTPFLCVSGDSFSVNPVRGNFLWRLFADK 537
H+ + EL + +R +L SL +K E + GDS S +PV+ N
Sbjct: 403 HQTAAYELQAFNLSWYWERIINLASLIKKGEQDRIASLKGDSKSSSPVKDNI-------S 455
Query: 536 FLKLDFRKPSTLVYVLASSKQVLK 465
LKL++ P L Y+ +S+Q +K
Sbjct: 456 RLKLEYHIPPILTYIGGASEQQMK 479
>gb|AAK16728.1|AF332962_1 ferrochelatase [Chlamydomonas reinhardtii]
Length = 493
Score = 35.8 bits (81), Expect = 0.91
Identities = 17/45 (37%), Positives = 26/45 (57%)
Frame = -3
Query: 743 VILFDDPDDHRRPSAELSGQTVDSLGKRTTSLNSLPRKDEGTPFL 609
V+ F DP+D R+P+A +G VD +G +L + D+ PFL
Sbjct: 64 VVSFVDPNDIRKPAAAAAGPAVDKVGVLLLNLGGPEKLDDVKPFL 108
>dbj|BAB63481.1| P0002B05.1 [Oryza sativa (japonica cultivar-group)]
gi|15528736|dbj|BAB64682.1| P0697C12.17 [Oryza sativa
(japonica cultivar-group)]
Length = 1043
Score = 35.0 bits (79), Expect = 1.6
Identities = 18/40 (45%), Positives = 21/40 (52%)
Frame = +3
Query: 783 DKDRRSTLRFSIGRRS*PEKAPDGGSGRGPRRRNDDPEHG 902
D DR S L +G + E+AP G G G RRR D P G
Sbjct: 289 DPDRVSILTIMLGVGASEERAPKGHDGAGERRRGDQPAPG 328
>ref|NP_253244.1| type 4 fimbrial biogenesis protein PilY1 [Pseudomonas aeruginosa
PA01] gi|11352777|pir||D83076 type 4 fimbrial biogenesis
protein PilY1 PA4554 [imported] - Pseudomonas aeruginosa
(strain PAO1) gi|9950799|gb|AAG07942.1|AE004869_6 type 4
fimbrial biogenesis protein PilY1 [Pseudomonas aeruginosa
PAO1]
Length = 1161
Score = 34.3 bits (77), Expect = 2.7
Identities = 31/121 (25%), Positives = 47/121 (38%), Gaps = 5/121 (4%)
Frame = -3
Query: 899 MFRIVVPSPRTAAAAAVGSFFRSTPTTNGKTECTAAILVARRVNPYFLFHISVILFDDPD 720
+ + + P+ A A + P T G+T T L + V D
Sbjct: 1052 LLQTITPNDDPCADGASNWTYGLDPYTGGRTSFTVFDLARQGV------------VDSKS 1099
Query: 719 DH--RRPSAELSGQTVDSLGKRTTSLNSLPRKDEGTPFLCVSGDSFSVNP---VRGNFLW 555
D+ + + +SG LG T S N ++G P +C SG+ +VNP RG W
Sbjct: 1100 DYSYNKQNVAVSGTEQKGLGGLTLSTN-----EQGNPEVCSSGECLTVNPGPNTRGRQNW 1154
Query: 554 R 552
R
Sbjct: 1155 R 1155
>pir||E71615 probable amine transporter PFB0435c - malaria parasite (Plasmodium
falciparum)
Length = 1138
Score = 33.9 bits (76), Expect = 3.5
Identities = 23/83 (27%), Positives = 45/83 (53%)
Frame = +2
Query: 287 KLTRSEHMLNYSSSYHSYIN**STYPLFDMKTIL*NK*QLLPNQLLNFNFHNNNENIYFS 466
K+ +SEH+ N + +N S Y + DM K + N+ N N++NN++N+ S
Sbjct: 209 KIVKSEHICNEKKT--KGVNGKSKYYIHDMN----RKDKQKKNKHNNINYNNNDDNVNNS 262
Query: 467 LKLAYSRLKHIQELKAYENQVLE 535
+ +S+ ++ Q + YEN++ +
Sbjct: 263 CEYNFSK-ENSQNMCHYENKIYD 284
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 731,295,437
Number of Sequences: 1393205
Number of extensions: 15786928
Number of successful extensions: 48765
Number of sequences better than 10.0: 23
Number of HSP's better than 10.0 without gapping: 46054
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 48713
length of database: 448,689,247
effective HSP length: 123
effective length of database: 277,325,032
effective search space used: 49363855696
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)