Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC005749A_C01 KMC005749A_c01
(609 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
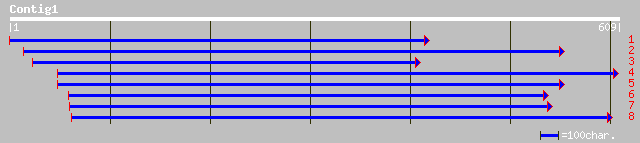
ref|NP_200265.1| putative protein; protein id: At5g54540.1, supp... 67 2e-10
ref|NP_682188.1| ORF_ID:tll1398~hypothetical protein [Thermosyne... 35 0.76
ref|NP_647905.1| CG15022-PA [Drosophila melanogaster] gi|2015208... 34 1.7
sp|Q12649|HMDH_PHYBL 3-hydroxy-3-methylglutaryl-coenzyme A reduc... 33 2.2
ref|NP_608926.2| CG6604-PA [Drosophila melanogaster] gi|7296977|... 33 2.2
>ref|NP_200265.1| putative protein; protein id: At5g54540.1, supported by cDNA:
gi_16648815, supported by cDNA: gi_20466128 [Arabidopsis
thaliana] gi|9758946|dbj|BAB09333.1|
gene_id:MRB17.4~pir||T05538~similar to unknown protein
[Arabidopsis thaliana] gi|16648816|gb|AAL25598.1|
AT5g54540/MRB17_4 [Arabidopsis thaliana]
gi|20466129|gb|AAM19986.1| AT5g54540/MRB17_4
[Arabidopsis thaliana]
Length = 297
Score = 67.0 bits (162), Expect = 2e-10
Identities = 46/142 (32%), Positives = 71/142 (49%)
Frame = -3
Query: 607 TVVASIASDSNVWKAVMQNPAVSSFFQSQQTVADFEAEGTTAKVVELTDDDTVTGSEEPE 428
TVVASIASD VW AVM+N + F Q+ +T + E + D + + E E
Sbjct: 170 TVVASIASDPKVWDAVMENKDLMKFLQTNKTAVSSQVES------DNDDQSERSSTTECE 223
Query: 427 IHSGIAFDFIGLLQNLKLTVAEMVSRVSNYLQNIFPTGEKEKSSGDADGNTKANVMDNMN 248
+ + + +LQ++KL ++ VS+Y ++F G S DG K + N
Sbjct: 224 VVETKPMELLEILQDMKLKAVRLMENVSSYFGDLFGLG-----SVTEDGKDKKQTLFN-- 276
Query: 247 VMGGSFMGLAMLVIMVVLVKRA 182
S GLA++VI +V++KRA
Sbjct: 277 -DPRSLFGLAVVVIFMVVLKRA 297
>ref|NP_682188.1| ORF_ID:tll1398~hypothetical protein [Thermosynechococcus elongatus
BP-1] gi|22295122|dbj|BAC08950.1|
ORF_ID:tll1398~hypothetical protein [Thermosynechococcus
elongatus BP-1]
Length = 340
Score = 35.0 bits (79), Expect = 0.76
Identities = 18/51 (35%), Positives = 24/51 (46%), Gaps = 5/51 (9%)
Frame = +2
Query: 110 QLI*LQSLRRLESST-----NCNLANQSLSSLHKYHHYHQHCQSHETSSHD 247
QL L R +E+ T NC L + H +HH+H H H + SHD
Sbjct: 262 QLFYLTRQREIETHTGQVAMNCETCKFRLVAGHGHHHHHPHDHEHHSHSHD 312
>ref|NP_647905.1| CG15022-PA [Drosophila melanogaster] gi|20152087|gb|AAM11403.1|
RE24507p [Drosophila melanogaster]
gi|23093010|gb|AAF47905.2| CG15022-PA [Drosophila
melanogaster]
Length = 295
Score = 33.9 bits (76), Expect = 1.7
Identities = 12/24 (50%), Positives = 14/24 (58%)
Frame = +2
Query: 191 HKYHHYHQHCQSHETSSHDIHIIH 262
H +HHY H H SSHD H +H
Sbjct: 46 HDHHHYDVHSHHHSPSSHDHHDLH 69
>sp|Q12649|HMDH_PHYBL 3-hydroxy-3-methylglutaryl-coenzyme A reductase (HMG-CoA reductase)
gi|9188117|emb|CAB97179.1| 3-hydroxy-3-methylglutaryl
coenzyme A reductase [Phycomyces blakesleeanus]
Length = 1176
Score = 33.5 bits (75), Expect = 2.2
Identities = 10/32 (31%), Positives = 18/32 (56%)
Frame = +2
Query: 191 HKYHHYHQHCQSHETSSHDIHIIHDIGFCISI 286
H +HH H H +H ++ H I+ I C+++
Sbjct: 705 HNHHHSHSHSHNHHSNHHQSDIVRPIDECVAL 736
>ref|NP_608926.2| CG6604-PA [Drosophila melanogaster] gi|7296977|gb|AAF52249.1|
CG6604-PA [Drosophila melanogaster]
Length = 660
Score = 33.5 bits (75), Expect = 2.2
Identities = 23/74 (31%), Positives = 34/74 (45%), Gaps = 1/74 (1%)
Frame = +2
Query: 74 QQTISYSVTPNVQLI*LQSLRRLESSTNCNLANQSLSSLHKYHHYHQH-CQSHETSSHDI 250
QQT S S T N + Q + ++ N+AN S H +H+H H +H +H+
Sbjct: 13 QQTPSPSQTQNFKSKLQQQIVSAAAAAAANIANGSSHHHHHQNHHHHHPLNNHHNHNHN- 71
Query: 251 HIIHDIGFCISISI 292
H+I F SI
Sbjct: 72 --QHNISFATDFSI 83
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 498,254,340
Number of Sequences: 1393205
Number of extensions: 10069947
Number of successful extensions: 38903
Number of sequences better than 10.0: 39
Number of HSP's better than 10.0 without gapping: 33265
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 37414
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 24283162270
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)