Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC005745A_C02 KMC005745A_c02
(1138 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
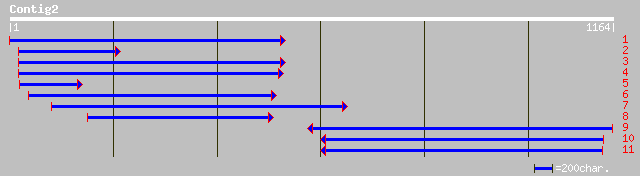
ref|NP_196231.2| putative protein; protein id: At5g06130.1, supp... 399 e-110
dbj|BAA98202.1| gene_id:K16F4.10~pir||T00468~similar to unknown ... 389 e-107
ref|NP_200975.1| putative protein; protein id: At5g61670.1, supp... 358 9e-98
ref|NP_181032.1| unknown protein; protein id: At2g34860.1, suppo... 46 0.001
ref|XP_144999.1| RIKEN cDNA D630042P16 gene [Mus musculus] 40 0.052
>ref|NP_196231.2| putative protein; protein id: At5g06130.1, supported by cDNA:
gi_18176019 [Arabidopsis thaliana]
gi|18176020|gb|AAL59969.1| unknown protein [Arabidopsis
thaliana] gi|22136730|gb|AAM91684.1| unknown protein
[Arabidopsis thaliana]
Length = 315
Score = 399 bits (1025), Expect = e-110
Identities = 199/261 (76%), Positives = 223/261 (85%)
Frame = -2
Query: 987 SSSKDPASSDNLPSNFCIIEGPETVQDFVQMQLQEIQGNIKSRRNKIFLLMEEVRRLRVQ 808
S+++ P S D +P+NFCIIEG ETVQDFVQMQLQEIQ NI+SRRNKIFLLMEEVRRLRVQ
Sbjct: 55 SNNRPPPSGDTVPNNFCIIEGSETVQDFVQMQLQEIQDNIRSRRNKIFLLMEEVRRLRVQ 114
Query: 807 QRLRGERRVISEEGEEEANEMPEIPSSIPFLPSVTPKTLKKLYLTSISFISAVIVFGGLI 628
QR++ + I+E+ E EA EMPEI SSIPFLP+VTPKTLK+LY TS++ IS +I FGGLI
Sbjct: 115 QRIKSVK-AINEDSELEATEMPEITSSIPFLPNVTPKTLKQLYSTSVALISGIIFFGGLI 173
Query: 627 APTLELKLGIGGTSYEDFIRSLHLPLQLSQVDPIVASFSGGAVGVISVLMLIEANNVKKQ 448
AP LELK+G+GGTSYEDFIRSLHLPLQLSQVDPIVASFSGGAVGVIS LMLIE NNVK+Q
Sbjct: 174 APNLELKVGLGGTSYEDFIRSLHLPLQLSQVDPIVASFSGGAVGVISTLMLIEVNNVKQQ 233
Query: 447 EKTMCKYCLGTGYLACARCSTSGVCLDIDPISVSSASVRPLQVPKTRRCPNCSGAGKVMC 268
EK CKYCLGTGYL CARCS SGVCL IDPI+ A+ + +QV T+RC NCSGAGKVMC
Sbjct: 234 EKKRCKYCLGTGYLPCARCSASGVCLSIDPITRPRATNQLMQVATTKRCLNCSGAGKVMC 293
Query: 267 PTCLCTGMKMASEHDLRIDPF 205
PTCLCTGM ASEHD R DPF
Sbjct: 294 PTCLCTGMVTASEHDPRFDPF 314
>dbj|BAA98202.1| gene_id:K16F4.10~pir||T00468~similar to unknown protein [Arabidopsis
thaliana]
Length = 319
Score = 389 bits (998), Expect = e-107
Identities = 210/309 (67%), Positives = 235/309 (75%), Gaps = 6/309 (1%)
Frame = -2
Query: 1113 SFPSPSKNST-FCSPFNGNTKQFSFSRGTTTLLQLQSRIILLRSSSKDPASSDNLP---- 949
S PS SK+ F S + + F + +LL S+ P SD L
Sbjct: 17 SSPSTSKSLLRFPSSYLKPSPSLLFHGSSRSLLSCSD------GSNNRPPPSDYLFGGYC 70
Query: 948 -SNFCIIEGPETVQDFVQMQLQEIQGNIKSRRNKIFLLMEEVRRLRVQQRLRGERRVISE 772
SNFCIIEG ETVQDFVQMQLQEIQ NI+SRRNKIFLLMEEVRRLRVQQR++ + I+E
Sbjct: 71 FSNFCIIEGSETVQDFVQMQLQEIQDNIRSRRNKIFLLMEEVRRLRVQQRIKSVK-AINE 129
Query: 771 EGEEEANEMPEIPSSIPFLPSVTPKTLKKLYLTSISFISAVIVFGGLIAPTLELKLGIGG 592
+ E EA EMPEI SSIPFLP+VTPKTLK+LY TS++ IS +I FGGLIAP LELK+G+GG
Sbjct: 130 DSELEATEMPEITSSIPFLPNVTPKTLKQLYSTSVALISGIIFFGGLIAPNLELKVGLGG 189
Query: 591 TSYEDFIRSLHLPLQLSQVDPIVASFSGGAVGVISVLMLIEANNVKKQEKTMCKYCLGTG 412
TSYEDFIRSLHLPLQLSQVDPIVASFSGGAVGVIS LMLIE NNVK+QEK CKYCLGTG
Sbjct: 190 TSYEDFIRSLHLPLQLSQVDPIVASFSGGAVGVISTLMLIEVNNVKQQEKKRCKYCLGTG 249
Query: 411 YLACARCSTSGVCLDIDPISVSSASVRPLQVPKTRRCPNCSGAGKVMCPTCLCTGMKMAS 232
YL CARCS SGVCL IDPI+ A+ + +QV T+RC NCSGAGKVMCPTCLCTGM AS
Sbjct: 250 YLPCARCSASGVCLSIDPITRPRATNQLMQVATTKRCLNCSGAGKVMCPTCLCTGMVTAS 309
Query: 231 EHDLRIDPF 205
EHD R DPF
Sbjct: 310 EHDPRFDPF 318
>ref|NP_200975.1| putative protein; protein id: At5g61670.1, supported by cDNA:
112078., supported by cDNA: gi_20453123 [Arabidopsis
thaliana] gi|9758482|dbj|BAB09011.1|
gene_id:K11J9.20~unknown protein [Arabidopsis thaliana]
gi|17529230|gb|AAL38842.1| unknown protein [Arabidopsis
thaliana] gi|20453124|gb|AAM19804.1| AT5g61670/k11j9_190
[Arabidopsis thaliana] gi|21436109|gb|AAM51301.1| unknown
protein [Arabidopsis thaliana] gi|21536802|gb|AAM61134.1|
unknown [Arabidopsis thaliana] gi|23506191|gb|AAN31107.1|
At5g61670/k11j9_190 [Arabidopsis thaliana]
Length = 307
Score = 358 bits (919), Expect = 9e-98
Identities = 188/294 (63%), Positives = 217/294 (72%), Gaps = 4/294 (1%)
Frame = -2
Query: 1074 PFNGNTKQFSFSRGTTTLLQLQSRIILL--RSSSKDPASS-DNLPSNFCIIEGPETVQDF 904
P+ Q+ S +L+ R L SSS D SS D S FCIIEGPETVQDF
Sbjct: 16 PYTWRFSQYKLSSSLGRNRRLRWRFTALDPESSSLDSESSADKFASGFCIIEGPETVQDF 75
Query: 903 VQMQLQEIQGNIKSRRNKIFLLMEEVRRLRVQQRLRG-ERRVISEEGEEEANEMPEIPSS 727
+MQLQEIQ NI+SRRNKIFL MEEVRRLR+QQR++ E +I+EE E +E+P PS
Sbjct: 76 AKMQLQEIQDNIRSRRNKIFLHMEEVRRLRIQQRIKNTELGIINEEQE---HELPNFPSF 132
Query: 726 IPFLPSVTPKTLKKLYLTSISFISAVIVFGGLIAPTLELKLGIGGTSYEDFIRSLHLPLQ 547
IPFLP +T LK Y T S I+ +I+FGGL+APTLELKLGIGGTSY DFI+SLHLP+Q
Sbjct: 133 IPFLPPLTAANLKVYYATCFSLIAGIILFGGLLAPTLELKLGIGGTSYADFIQSLHLPMQ 192
Query: 546 LSQVDPIVASFSGGAVGVISVLMLIEANNVKKQEKTMCKYCLGTGYLACARCSTSGVCLD 367
LSQVDPIVASFSGGAVGVIS LM++E NNVK+QE CKYCLGTGYLACARCS++G +
Sbjct: 193 LSQVDPIVASFSGGAVGVISALMVVEVNNVKQQEHKRCKYCLGTGYLACARCSSTGALVL 252
Query: 366 IDPISVSSASVRPLQVPKTRRCPNCSGAGKVMCPTCLCTGMKMASEHDLRIDPF 205
+P+S + L PKT RC NCSGAGKVMCPTCLCTGM MASEHD RIDPF
Sbjct: 253 TEPVSAIAGGNHSLSPPKTERCSNCSGAGKVMCPTCLCTGMAMASEHDPRIDPF 306
>ref|NP_181032.1| unknown protein; protein id: At2g34860.1, supported by cDNA:
gi_20466395 [Arabidopsis thaliana]
gi|7485815|pir||T00468 hypothetical protein At2g34860
[imported] - Arabidopsis thaliana
gi|3033382|gb|AAC12826.1| unknown protein [Arabidopsis
thaliana] gi|20466396|gb|AAM20515.1| unknown protein
[Arabidopsis thaliana] gi|22136346|gb|AAM91251.1|
unknown protein [Arabidopsis thaliana]
Length = 186
Score = 46.2 bits (108), Expect = 0.001
Identities = 22/64 (34%), Positives = 32/64 (49%)
Frame = -2
Query: 435 CKYCLGTGYLACARCSTSGVCLDIDPISVSSASVRPLQVPKTRRCPNCSGAGKVMCPTCL 256
C+ C G+G + C C +G ++ R V + CPNC G GK++CP CL
Sbjct: 102 CRNCQGSGAVLCDMCGGTGKWKALN-------RKRAKDVYEFTECPNCYGRGKLVCPVCL 154
Query: 255 CTGM 244
TG+
Sbjct: 155 GTGL 158
>ref|XP_144999.1| RIKEN cDNA D630042P16 gene [Mus musculus]
Length = 438
Score = 40.4 bits (93), Expect = 0.052
Identities = 22/65 (33%), Positives = 30/65 (45%)
Frame = -2
Query: 435 CKYCLGTGYLACARCSTSGVCLDIDPISVSSASVRPLQVPKTRRCPNCSGAGKVMCPTCL 256
C C G G C+ C +G+ + SS S + + RRC CSG+G+ C TC
Sbjct: 246 CHKCHGRGRYKCSGCHGAGM------VRCSSCSGTKRKAKQPRRCHLCSGSGRRRCSTCS 299
Query: 255 CTGMK 241
G K
Sbjct: 300 GRGNK 304
Score = 35.0 bits (79), Expect = 2.2
Identities = 23/77 (29%), Positives = 30/77 (38%), Gaps = 1/77 (1%)
Frame = -2
Query: 486 VLMLIEANNVKKQEKTMCKYCLGTGYLACARCS-TSGVCLDIDPISVSSASVRPLQVPKT 310
+L E + + + C C G G + C+ CS T + S S R
Sbjct: 240 ILAFQECHKCHGRGRYKCSGCHGAGMVRCSSCSGTKRKAKQPRRCHLCSGSGR------- 292
Query: 309 RRCPNCSGAGKVMCPTC 259
RRC CSG G C TC
Sbjct: 293 RRCSTCSGRGNKTCATC 309
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 993,329,318
Number of Sequences: 1393205
Number of extensions: 22847970
Number of successful extensions: 94723
Number of sequences better than 10.0: 64
Number of HSP's better than 10.0 without gapping: 72367
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 91373
length of database: 448,689,247
effective HSP length: 125
effective length of database: 274,538,622
effective search space used: 69458271366
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)