Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC005741A_C01 KMC005741A_c01
(623 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
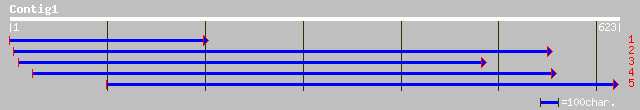
gb|AAF64170.1|AF242547_1 plastid ribosomal protein S9 precursor ... 208 4e-53
ref|NP_177635.1| ribosomal protein S9, putative; protein id: At1... 207 7e-53
gb|AAK16543.1|AF332374_1 9S ribosomal protein [Zea mays] gi|1324... 187 1e-46
dbj|BAA82395.1| ribosomal protein S9 [Oryza sativa (japonica cul... 184 8e-46
pir||E96779 probable ribosomal protein S9 F9E10.18 [imported] - ... 174 1e-42
>gb|AAF64170.1|AF242547_1 plastid ribosomal protein S9 precursor [Spinacia oleracea]
Length = 197
Score = 208 bits (530), Expect = 4e-53
Identities = 102/114 (89%), Positives = 107/114 (93%)
Frame = -1
Query: 623 RVVLQEGTGKFIINYRDAKEYLQGNPLWLQYIKVPLVTLGYETSYDVFVKAEGGGLSGQA 444
RVVLQEGTGKFIINYRDAKEYLQGNPLWLQY+K PL TLGYET+YDVFVKA GGGLSGQA
Sbjct: 84 RVVLQEGTGKFIINYRDAKEYLQGNPLWLQYVKTPLATLGYETNYDVFVKAHGGGLSGQA 143
Query: 443 QAISLGIARALLKVSEDHRVPLRQEGLLTRDSRVVERKKAGLKKARKAPQFSKR 282
QAISLG+ARALLKVS HR PL+QEGLLTRDSR+VERKK GLKKARKAPQFSKR
Sbjct: 144 QAISLGVARALLKVSASHRAPLKQEGLLTRDSRIVERKKPGLKKARKAPQFSKR 197
>ref|NP_177635.1| ribosomal protein S9, putative; protein id: At1g74970.1, supported
by cDNA: 250383., supported by cDNA: gi_12744978,
supported by cDNA: gi_15010597, supported by cDNA:
gi_20259210, supported by cDNA: gi_5456945 [Arabidopsis
thaliana] gi|25456470|pir||T52450 ribosomal protein S9
[imported] - Arabidopsis thaliana
gi|5456946|dbj|BAA82396.1| ribosomal protein S9
[Arabidopsis thaliana]
gi|5882726|gb|AAD55279.1|AC008263_10 Identical to
gb|AB022676 ribosomal protein S9 from Arabidopsis
thaliana. ESTs gb|T13861, gb|AA389790, gb|T42539,
gb|AA586013, gb|AA395093 and gb|AA041154 come from this
gene gi|12744979|gb|AAK06869.1|AF344318_1 putative
ribosomal protein S9 [Arabidopsis thaliana]
gi|15010598|gb|AAK73958.1| ATg74970/F25A4.6 [Arabidopsis
thaliana] gi|15027999|gb|AAK76530.1| putative ribosomal
protein S9 [Arabidopsis thaliana]
gi|20259211|gb|AAM14321.1| putative ribosomal protein S9
[Arabidopsis thaliana] gi|21554316|gb|AAM63421.1|
ribosomal protein S9, putative [Arabidopsis thaliana]
Length = 208
Score = 207 bits (528), Expect = 7e-53
Identities = 102/114 (89%), Positives = 109/114 (95%)
Frame = -1
Query: 623 RVVLQEGTGKFIINYRDAKEYLQGNPLWLQYIKVPLVTLGYETSYDVFVKAEGGGLSGQA 444
RVVLQEGTGK IINYRDAKEYLQGNPLWLQY+KVPLVTLGYE SYD+FVKA GGGLSGQA
Sbjct: 95 RVVLQEGTGKVIINYRDAKEYLQGNPLWLQYVKVPLVTLGYENSYDIFVKAHGGGLSGQA 154
Query: 443 QAISLGIARALLKVSEDHRVPLRQEGLLTRDSRVVERKKAGLKKARKAPQFSKR 282
QAI+LG+ARALLKVS DHR PL++EGLLTRD+RVVERKKAGLKKARKAPQFSKR
Sbjct: 155 QAITLGVARALLKVSADHRSPLKKEGLLTRDARVVERKKAGLKKARKAPQFSKR 208
>gb|AAK16543.1|AF332374_1 9S ribosomal protein [Zea mays]
gi|13242066|gb|AAK16544.1|AF332375_1 9S ribosomal
protein [Zea mays]
Length = 221
Score = 187 bits (474), Expect = 1e-46
Identities = 89/114 (78%), Positives = 103/114 (90%)
Frame = -1
Query: 623 RVVLQEGTGKFIINYRDAKEYLQGNPLWLQYIKVPLVTLGYETSYDVFVKAEGGGLSGQA 444
RVVLQEGTGK IN+RDAKEYLQGNP+W++Y KVPL+TLG+E +YDVFVK GGGLSGQA
Sbjct: 108 RVVLQEGTGKVFINFRDAKEYLQGNPMWMEYCKVPLMTLGFEKNYDVFVKVHGGGLSGQA 167
Query: 443 QAISLGIARALLKVSEDHRVPLRQEGLLTRDSRVVERKKAGLKKARKAPQFSKR 282
QAI LG+ARAL+K+S +RVPL+ EGLLTRD+R+VERKKAGLKKARK PQFSKR
Sbjct: 168 QAICLGVARALVKISNANRVPLKSEGLLTRDTRIVERKKAGLKKARKRPQFSKR 221
>dbj|BAA82395.1| ribosomal protein S9 [Oryza sativa (japonica cultivar-group)]
Length = 223
Score = 184 bits (467), Expect = 8e-46
Identities = 89/114 (78%), Positives = 102/114 (89%)
Frame = -1
Query: 623 RVVLQEGTGKFIINYRDAKEYLQGNPLWLQYIKVPLVTLGYETSYDVFVKAEGGGLSGQA 444
RVVLQEGTG+ IN+RDAKEYLQGNP+W++Y KVPLVTLG+E SYDVFVK GGGLSGQA
Sbjct: 110 RVVLQEGTGRVFINFRDAKEYLQGNPMWMEYCKVPLVTLGFENSYDVFVKVHGGGLSGQA 169
Query: 443 QAISLGIARALLKVSEDHRVPLRQEGLLTRDSRVVERKKAGLKKARKAPQFSKR 282
QAI LG+ARAL+K+S ++V LR EGLLTRD+R+VERKKAGLKKARK PQFSKR
Sbjct: 170 QAICLGVARALVKISTANKVTLRGEGLLTRDTRIVERKKAGLKKARKRPQFSKR 223
>pir||E96779 probable ribosomal protein S9 F9E10.18 [imported] - Arabidopsis
thaliana gi|12323890|gb|AAG51916.1|AC013258_10 putative
ribosomal protein S9; 45292-45606 [Arabidopsis thaliana]
Length = 104
Score = 174 bits (440), Expect = 1e-42
Identities = 84/96 (87%), Positives = 92/96 (95%)
Frame = -1
Query: 569 KEYLQGNPLWLQYIKVPLVTLGYETSYDVFVKAEGGGLSGQAQAISLGIARALLKVSEDH 390
+EYLQGNPLWLQY+KVPLVTLGYE SYD+FVKA GGGLSGQAQAI+LG+ARALLKVS DH
Sbjct: 9 QEYLQGNPLWLQYVKVPLVTLGYENSYDIFVKAHGGGLSGQAQAITLGVARALLKVSADH 68
Query: 389 RVPLRQEGLLTRDSRVVERKKAGLKKARKAPQFSKR 282
R PL++EGLLTRD+RVVERKKAGLKKARKAPQFSKR
Sbjct: 69 RSPLKKEGLLTRDARVVERKKAGLKKARKAPQFSKR 104
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 534,029,827
Number of Sequences: 1393205
Number of extensions: 11280082
Number of successful extensions: 26595
Number of sequences better than 10.0: 198
Number of HSP's better than 10.0 without gapping: 25947
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 26521
length of database: 448,689,247
effective HSP length: 118
effective length of database: 284,291,057
effective search space used: 25301904073
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)