Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC005716A_C01 KMC005716A_c01
(580 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
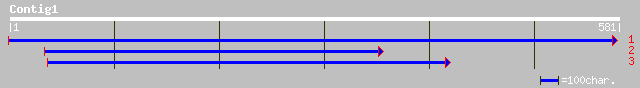
dbj|BAB90707.1| putative cdc21 protein [Oryza sativa (japonica c... 144 8e-34
ref|NP_179236.1| putative CDC21 protein; protein id: At2g16440.1... 116 2e-25
dbj|BAC41036.1| unnamed protein product [Mus musculus] 79 4e-14
dbj|BAB27813.1| unnamed protein product [Mus musculus] 79 4e-14
ref|NP_032591.1| mini chromosome maintenance deficient 4 homolog... 79 4e-14
>dbj|BAB90707.1| putative cdc21 protein [Oryza sativa (japonica cultivar-group)]
Length = 849
Score = 144 bits (363), Expect = 8e-34
Identities = 71/99 (71%), Positives = 90/99 (90%)
Frame = -2
Query: 579 ARIRFSELVEKRDVMEAFRLLEVAMQQSATDHSTGTIDMDLITTGVSASERMRRESLLQA 400
AR+RFSE+VE +DV+EAFRLLEVAMQQSATDH+TGTIDMDLI TG+SASER RR++L+ A
Sbjct: 742 ARMRFSEMVEVQDVVEAFRLLEVAMQQSATDHATGTIDMDLIMTGISASERQRRDNLVAA 801
Query: 399 TRNIIMEKMQIGGPSKRLLELLEELKQQNPGSEIHLADV 283
TRN++MEKMQ+GGPS R++ELLEE+++Q+ E+HL DV
Sbjct: 802 TRNLVMEKMQLGGPSVRMIELLEEIRKQS-SMEVHLHDV 839
>ref|NP_179236.1| putative CDC21 protein; protein id: At2g16440.1 [Arabidopsis
thaliana] gi|25411709|pir||C84540 probable CDC21 protein
[imported] - Arabidopsis thaliana
gi|4544386|gb|AAD22296.1| putative CDC21 protein
[Arabidopsis thaliana]
Length = 720
Score = 116 bits (290), Expect = 2e-25
Identities = 66/101 (65%), Positives = 78/101 (76%), Gaps = 2/101 (1%)
Frame = -2
Query: 579 ARIRFSELVEKRDVMEAFRLLEVAMQQSATDHSTGTIDMDLITTGVSASERMRRESLLQA 400
AR+RFSE VEK DV EAFRLL VAMQQSATDH+TGTIDMDLI TGVSASERMRR++ +
Sbjct: 620 ARMRFSEWVEKHDVDEAFRLLRVAMQQSATDHATGTIDMDLINTGVSASERMRRDTFASS 679
Query: 399 TRNIIMEKMQIGGPSKRLLELLEELKQQ--NPGSEIHLADV 283
R+I +EK+ + LL+LLEELK+ N +EIHL DV
Sbjct: 680 IRDIALEKITM---EMLLLQLLEELKKHGGNINTEIHLHDV 717
>dbj|BAC41036.1| unnamed protein product [Mus musculus]
Length = 862
Score = 79.0 bits (193), Expect = 4e-14
Identities = 46/120 (38%), Positives = 75/120 (62%)
Frame = -2
Query: 579 ARIRFSELVEKRDVMEAFRLLEVAMQQSATDHSTGTIDMDLITTGVSASERMRRESLLQA 400
A++RFS VE DV EA RL A++QSATD TG +D+ ++TTG+SA+ R R+E L +A
Sbjct: 744 AKVRFSNKVEAIDVEEAKRLHREALKQSATDPRTGMVDISILTTGMSATSRKRKEELAEA 803
Query: 399 TRNIIMEKMQIGGPSKRLLELLEELKQQNPGSEIHLADVRTAIATLASEGFITMHGDSVK 220
R +I+ K + P+ + +L E+++ Q+ + I A+ LA + F+T+ G +V+
Sbjct: 804 LRKLILSKGKT--PALKYQQLFEDIRGQS-DTAITKDMFEEALRALADDDFLTVTGKTVR 860
>dbj|BAB27813.1| unnamed protein product [Mus musculus]
Length = 862
Score = 79.0 bits (193), Expect = 4e-14
Identities = 46/120 (38%), Positives = 75/120 (62%)
Frame = -2
Query: 579 ARIRFSELVEKRDVMEAFRLLEVAMQQSATDHSTGTIDMDLITTGVSASERMRRESLLQA 400
A++RFS VE DV EA RL A++QSATD TG +D+ ++TTG+SA+ R R+E L +A
Sbjct: 744 AKVRFSNKVEAIDVEEAKRLHREALKQSATDPRTGIVDISILTTGMSATSRKRKEELAEA 803
Query: 399 TRNIIMEKMQIGGPSKRLLELLEELKQQNPGSEIHLADVRTAIATLASEGFITMHGDSVK 220
R +I+ K + P+ + +L E+++ Q+ + I A+ LA + F+T+ G +V+
Sbjct: 804 LRKLILSKGKT--PALKYQQLFEDIRGQS-DTAITKDMFEEALRALADDDFLTVTGKTVR 860
>ref|NP_032591.1| mini chromosome maintenance deficient 4 homolog (S. cerevisiae) [Mus
musculus] gi|1705521|sp|P49717|MCM4_MOUSE DNA REPLICATION
LICENSING FACTOR MCM4 (CDC21 HOMOLOG) (P1-CDC21)
gi|1363105|pir||S56766 replication licensing factor MCM4
- mouse gi|940406|dbj|BAA05082.1| mcdc21 protein [Mus
musculus] gi|26353896|dbj|BAC40578.1| unnamed protein
product [Mus musculus]
Length = 862
Score = 79.0 bits (193), Expect = 4e-14
Identities = 46/120 (38%), Positives = 75/120 (62%)
Frame = -2
Query: 579 ARIRFSELVEKRDVMEAFRLLEVAMQQSATDHSTGTIDMDLITTGVSASERMRRESLLQA 400
A++RFS VE DV EA RL A++QSATD TG +D+ ++TTG+SA+ R R+E L +A
Sbjct: 744 AKVRFSNKVEAIDVEEAKRLHREALKQSATDPRTGIVDISILTTGMSATSRKRKEELAEA 803
Query: 399 TRNIIMEKMQIGGPSKRLLELLEELKQQNPGSEIHLADVRTAIATLASEGFITMHGDSVK 220
R +I+ K + P+ + +L E+++ Q+ + I A+ LA + F+T+ G +V+
Sbjct: 804 LRKLILSKGKT--PALKYQQLFEDIRGQS-DTAITKDMFEEALRALADDDFLTVTGKTVR 860
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 500,006,993
Number of Sequences: 1393205
Number of extensions: 10298106
Number of successful extensions: 26255
Number of sequences better than 10.0: 52
Number of HSP's better than 10.0 without gapping: 25519
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 26244
length of database: 448,689,247
effective HSP length: 117
effective length of database: 285,684,262
effective search space used: 21426319650
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)