Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
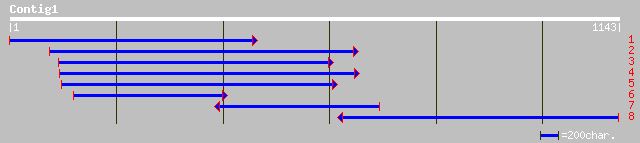
Query= KMC005699A_C01 KMC005699A_c01
(1136 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|NP_084748.1| hypothetical protein [Oenothera elata subsp. ho... 120 4e-26
ref|NP_759470.1| Unknown [Vibrio vulnificus CMCP6] gi|27364365|r... 70 4e-13
ref|NP_760124.1| Unknown [Vibrio vulnificus CMCP6] gi|27364802|r... 69 8e-13
ref|NP_616552.1| predicted protein [Methanosarcina acetivorans s... 44 2e-04
ref|NP_799509.1| hypothetical protein [Bacillus megaterium] gi|2... 39 0.001
>ref|NP_084748.1| hypothetical protein [Oenothera elata subsp. hookeri]
gi|13518401|ref|NP_084760.1| hypothetical protein
[Oenothera elata subsp. hookeri]
gi|6723805|emb|CAB67216.1| hypothetical protein
[Oenothera elata subsp. hookeri]
gi|6723818|emb|CAB67229.1| hypothetical protein
[Oenothera elata subsp. hookeri]
Length = 54
Score = 120 bits (301), Expect = 4e-26
Identities = 52/54 (96%), Positives = 53/54 (97%)
Frame = -1
Query: 338 IALPQPRFHGLGCSHFARRYYGNRFCFLFLWLLRCFSSPGCLLPAHGFSSSSKG 177
+ LPQPRFHGLGCSHFARRYYGNRFCFLFLWLLRCFSSPGCLLPAHGFSSSSKG
Sbjct: 1 MTLPQPRFHGLGCSHFARRYYGNRFCFLFLWLLRCFSSPGCLLPAHGFSSSSKG 54
>ref|NP_759470.1| Unknown [Vibrio vulnificus CMCP6] gi|27364365|ref|NP_759893.1|
Unknown [Vibrio vulnificus CMCP6]
gi|27364371|ref|NP_759899.1| Unknown [Vibrio vulnificus
CMCP6] gi|27364410|ref|NP_759938.1| Unknown [Vibrio
vulnificus CMCP6] gi|27364494|ref|NP_760022.1| Unknown
[Vibrio vulnificus CMCP6] gi|27364876|ref|NP_760404.1|
Unknown [Vibrio vulnificus CMCP6]
gi|27360059|gb|AAO08997.1|AE016798_157 Unknown [Vibrio
vulnificus CMCP6] gi|27360484|gb|AAO09420.1|AE016800_25
Unknown [Vibrio vulnificus CMCP6]
gi|27360490|gb|AAO09426.1|AE016800_31 Unknown [Vibrio
vulnificus CMCP6] gi|27360529|gb|AAO09465.1|AE016800_70
Unknown [Vibrio vulnificus CMCP6]
gi|27360613|gb|AAO09549.1|AE016800_154 Unknown [Vibrio
vulnificus CMCP6] gi|27361021|gb|AAO09931.1|AE016801_250
Unknown [Vibrio vulnificus CMCP6]
Length = 105
Score = 70.5 bits (171), Expect(2) = 4e-13
Identities = 38/59 (64%), Positives = 43/59 (72%), Gaps = 1/59 (1%)
Frame = -2
Query: 559 PRWGFFSPFPHGTTSLSVTQEYLALQGGPC*FTRDSTCPMLLG-SEHKLVMLSATGLSP 386
P GFFSPFPHGT SLSV+QEYLAL+ GP F +D+TCP LL +EH S TGLSP
Sbjct: 40 PSQGFFSPFPHGTGSLSVSQEYLALEDGPPIFRQDNTCPALLDFTEH---ASSTTGLSP 95
Score = 27.3 bits (59), Expect(2) = 4e-13
Identities = 14/22 (63%), Positives = 14/22 (63%)
Frame = -3
Query: 600 PTAWELTVSCSISLPDGGSFHP 535
PTA TVS SISLP G F P
Sbjct: 26 PTACTYTVSGSISLPSQGFFSP 47
>ref|NP_760124.1| Unknown [Vibrio vulnificus CMCP6] gi|27364802|ref|NP_760330.1|
Unknown [Vibrio vulnificus CMCP6]
gi|27367802|ref|NP_763329.1| Unknown [Vibrio vulnificus
CMCP6] gi|27359375|gb|AAO08319.1|AE016813_71 Unknown
[Vibrio vulnificus CMCP6]
gi|27360715|gb|AAO09651.1|AE016800_256 Unknown [Vibrio
vulnificus CMCP6] gi|27360947|gb|AAO09857.1|AE016801_176
Unknown [Vibrio vulnificus CMCP6]
Length = 105
Score = 69.3 bits (168), Expect(2) = 8e-13
Identities = 37/59 (62%), Positives = 43/59 (72%), Gaps = 1/59 (1%)
Frame = -2
Query: 559 PRWGFFSPFPHGTTSLSVTQEYLALQGGPC*FTRDSTCPMLLG-SEHKLVMLSATGLSP 386
P GFFSPFPHGT SLSV+QEYLAL+ GP F +D+TCP LL +EH + TGLSP
Sbjct: 40 PSQGFFSPFPHGTGSLSVSQEYLALEDGPPIFRQDNTCPALLDFTEH---ASTTTGLSP 95
Score = 27.3 bits (59), Expect(2) = 8e-13
Identities = 14/22 (63%), Positives = 14/22 (63%)
Frame = -3
Query: 600 PTAWELTVSCSISLPDGGSFHP 535
PTA TVS SISLP G F P
Sbjct: 26 PTACTYTVSGSISLPSQGFFSP 47
>ref|NP_616552.1| predicted protein [Methanosarcina acetivorans str. C2A]
gi|19915496|gb|AAM05032.1| predicted protein
[Methanosarcina acetivorans str. C2A]
Length = 125
Score = 44.3 bits (103), Expect(2) = 2e-04
Identities = 27/46 (58%), Positives = 30/46 (64%), Gaps = 4/46 (8%)
Frame = -3
Query: 351 ARRLYSSPTTPFS----RFRLLPFRSPLLRESLLLSFPLATKMFQF 226
ARRL +SPTTP RF L F SPLL S L+SFP T+MFQF
Sbjct: 44 ARRL-NSPTTPHVPEGIRFELCRFHSPLLTASQLISFPAPTEMFQF 88
Score = 24.3 bits (51), Expect(2) = 2e-04
Identities = 11/23 (47%), Positives = 12/23 (51%)
Frame = -2
Query: 169 FGNLRIYAYFQLPEAFRRLLRPS 101
FGN LP A+R L RPS
Sbjct: 103 FGNPGFIGSMHLPRAYRSLARPS 125
>ref|NP_799509.1| hypothetical protein [Bacillus megaterium]
gi|28848606|gb|AAO52806.1| hypothetical protein
[Bacillus megaterium]
Length = 79
Score = 38.5 bits (88), Expect(2) = 0.001
Identities = 23/37 (62%), Positives = 25/37 (67%)
Frame = -2
Query: 304 AAPISLAATTGIAFAFFSSGY*DVSVRQVVSCLPMDS 194
A +SLAAT IAFAF SSGY DVSV +V PM S
Sbjct: 4 ANSVSLAATQEIAFAFSSSGYLDVSVPRVCLQYPMYS 40
Score = 27.3 bits (59), Expect(2) = 0.001
Identities = 14/29 (48%), Positives = 17/29 (58%)
Frame = -1
Query: 179 G*PIRESPDLCLFSTPRSISSLTTPLLVS 93
G PIR+SPD + PRSIS L + S
Sbjct: 51 GFPIRKSPDQSSLTAPRSISVLVPSFIGS 79
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,037,691,134
Number of Sequences: 1393205
Number of extensions: 24916481
Number of successful extensions: 68453
Number of sequences better than 10.0: 24
Number of HSP's better than 10.0 without gapping: 64457
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 68417
length of database: 448,689,247
effective HSP length: 125
effective length of database: 274,538,622
effective search space used: 69458271366
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)