Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
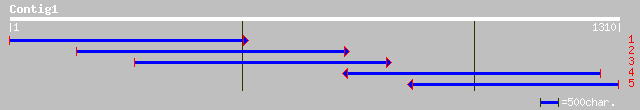
Query= KMC005698A_C01 KMC005698A_c01
(1271 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
sp|O82043|ILV5_PEA Ketol-acid reductoisomerase, chloroplast prec... 578 e-164
emb|CAB61890.1| acetohydroxy acid isomeroreductase [Pisum sativum] 577 e-163
ref|NP_191420.1| ketol-acid reductoisomerase; protein id: At3g58... 562 e-159
pir||S30145 ketol-acid reductoisomerase (EC 1.1.1.86) precursor ... 560 e-158
emb|CAA48253.1| ketol-acid reductoisomerase [Arabidopsis thaliana] 560 e-158
>sp|O82043|ILV5_PEA Ketol-acid reductoisomerase, chloroplast precursor (Acetohydroxy-acid
reductoisomerase) (Alpha-keto-beta-hydroxylacil
reductoisomerase) gi|7431346|pir||T06825 ketol-acid
reductoisomerase (EC 1.1.1.86) - garden pea
gi|3452497|emb|CAA76854.1| ketol-acid reductoisomerase
[Pisum sativum]
Length = 581
Score = 578 bits (1489), Expect = e-164
Identities = 286/304 (94%), Positives = 298/304 (97%)
Frame = +2
Query: 2 WSVALGSPFTFATTLEQEYKSDIFGERGILLGAVHGIVESLFRRYTENGMAEDLAYKNTV 181
WSVALGSPFTFATTLEQEYKSDIFGERGILLGAVHGIVESLFRRYTENGM+EDLAYKNTV
Sbjct: 278 WSVALGSPFTFATTLEQEYKSDIFGERGILLGAVHGIVESLFRRYTENGMSEDLAYKNTV 337
Query: 182 ESITGIISKTISTKGMLAVYNSFSEEGKREFEKAYSASFYPCMDILYECYEDVASGSEIR 361
ESITG+ISKTIST+GMLAVYN+ SE+GK+EFEKAYSASFYPCM+ILYECYEDVASGSEIR
Sbjct: 338 ESITGVISKTISTQGMLAVYNALSEDGKKEFEKAYSASFYPCMEILYECYEDVASGSEIR 397
Query: 362 SVVLAGRRFYEKEGLPAFPMGKIDQTRMWKVGERVRSTRPAGDLGPLNPFTAGVFVALMM 541
SVVLAGRRFYEKEGLPAFPMGKIDQTRMWKVGERVRSTRPAGDLGPL PFTAGVFVA+MM
Sbjct: 398 SVVLAGRRFYEKEGLPAFPMGKIDQTRMWKVGERVRSTRPAGDLGPLYPFTAGVFVAMMM 457
Query: 542 AQIEVLRKKGHSYSEIINESVIESVDSLNPFMHARGVSFMVDNCSTTARLGSRKWAPRFD 721
AQIEVLRKKGHSYSEIINESVIESVDSLNPFMHARGVSFMVDNCSTTARLGSRKWAPRFD
Sbjct: 458 AQIEVLRKKGHSYSEIINESVIESVDSLNPFMHARGVSFMVDNCSTTARLGSRKWAPRFD 517
Query: 722 YVLTQQALVAVDNAAPINQDLISNFLSDPVHGAIEVCAELRPTLDISVPPDADFVRPELR 901
Y+LTQQALVAVD+ APINQDLISNF+SDPVHGAI+VCAELRPTLDISVP ADFVRPELR
Sbjct: 518 YILTQQALVAVDSGAPINQDLISNFVSDPVHGAIQVCAELRPTLDISVPAAADFVRPELR 577
Query: 902 QSSN 913
Q SN
Sbjct: 578 QCSN 581
>emb|CAB61890.1| acetohydroxy acid isomeroreductase [Pisum sativum]
Length = 581
Score = 577 bits (1486), Expect = e-163
Identities = 285/304 (93%), Positives = 298/304 (97%)
Frame = +2
Query: 2 WSVALGSPFTFATTLEQEYKSDIFGERGILLGAVHGIVESLFRRYTENGMAEDLAYKNTV 181
WSVALGSPFTFATTLEQEYKSDIFGERGILLGAVHGIVESLFRRYTENGM+EDLAYKNTV
Sbjct: 278 WSVALGSPFTFATTLEQEYKSDIFGERGILLGAVHGIVESLFRRYTENGMSEDLAYKNTV 337
Query: 182 ESITGIISKTISTKGMLAVYNSFSEEGKREFEKAYSASFYPCMDILYECYEDVASGSEIR 361
ESITG+ISKTIST+GMLAVYN+ SE+GK+EFE+AYSASFYPCM+ILYECYEDVASGSEIR
Sbjct: 338 ESITGVISKTISTQGMLAVYNALSEDGKKEFERAYSASFYPCMEILYECYEDVASGSEIR 397
Query: 362 SVVLAGRRFYEKEGLPAFPMGKIDQTRMWKVGERVRSTRPAGDLGPLNPFTAGVFVALMM 541
SVVLAGRRFYEKEGLPAFPMGKIDQTRMWKVGERVRSTRPAGDLGPL PFTAGVFVA+MM
Sbjct: 398 SVVLAGRRFYEKEGLPAFPMGKIDQTRMWKVGERVRSTRPAGDLGPLYPFTAGVFVAMMM 457
Query: 542 AQIEVLRKKGHSYSEIINESVIESVDSLNPFMHARGVSFMVDNCSTTARLGSRKWAPRFD 721
AQIEVLRKKGHSYSEIINESVIESVDSLNPFMHARGVSFMVDNCSTTARLGSRKWAPRFD
Sbjct: 458 AQIEVLRKKGHSYSEIINESVIESVDSLNPFMHARGVSFMVDNCSTTARLGSRKWAPRFD 517
Query: 722 YVLTQQALVAVDNAAPINQDLISNFLSDPVHGAIEVCAELRPTLDISVPPDADFVRPELR 901
Y+LTQQALVAVD+ APINQDLISNF+SDPVHGAI+VCAELRPTLDISVP ADFVRPELR
Sbjct: 518 YILTQQALVAVDSGAPINQDLISNFVSDPVHGAIQVCAELRPTLDISVPAAADFVRPELR 577
Query: 902 QSSN 913
Q SN
Sbjct: 578 QCSN 581
>ref|NP_191420.1| ketol-acid reductoisomerase; protein id: At3g58610.1, supported by
cDNA: gi_11993866, supported by cDNA: gi_13194831,
supported by cDNA: gi_13265432, supported by cDNA:
gi_17063194, supported by cDNA: gi_17529223, supported by
cDNA: gi_20465492 [Arabidopsis thaliana]
gi|12644387|sp|Q05758|ILV5_ARATH Ketol-acid
reductoisomerase, chloroplast precursor
(Acetohydroxy-acid reductoisomerase)
(Alpha-keto-beta-hydroxylacil reductoisomerase)
gi|11251579|pir||T45681 ketol-acid reductoisomerase -
Arabidopsis thaliana gi|402552|emb|CAA49506.1| ketol-acid
reductoisomerase [Arabidopsis thaliana]
gi|6735378|emb|CAB68199.1| ketol-acid reductoisomerase
[Arabidopsis thaliana]
gi|11692838|gb|AAG40022.1|AF324671_1 AT3g58610
[Arabidopsis thaliana]
gi|11993867|gb|AAG42917.1|AF329500_1 putative ketol-acid
reductoisomerase [Arabidopsis thaliana]
gi|17063195|gb|AAL32973.1| AT3g58610/F14P22_200
[Arabidopsis thaliana] gi|17529224|gb|AAL38839.1|
putative ketol-acid reductoisomerase [Arabidopsis
thaliana] gi|20465493|gb|AAM20206.1| putative ketol-acid
reductoisomerase [Arabidopsis thaliana]
gi|23397061|gb|AAN31816.1| putative ketol-acid
reductoisomerase [Arabidopsis thaliana]
gi|23463055|gb|AAN33197.1| At3g58610/F14P22_200
[Arabidopsis thaliana]
Length = 591
Score = 562 bits (1448), Expect = e-159
Identities = 277/304 (91%), Positives = 292/304 (95%)
Frame = +2
Query: 2 WSVALGSPFTFATTLEQEYKSDIFGERGILLGAVHGIVESLFRRYTENGMAEDLAYKNTV 181
WSVALGSPFTFATTLEQEY+SDIFGERGILLGAVHGIVESLFRRYTENGM+EDLAYKNTV
Sbjct: 288 WSVALGSPFTFATTLEQEYRSDIFGERGILLGAVHGIVESLFRRYTENGMSEDLAYKNTV 347
Query: 182 ESITGIISKTISTKGMLAVYNSFSEEGKREFEKAYSASFYPCMDILYECYEDVASGSEIR 361
E ITG IS+TIST+GMLAVYNS SEEGK++FE AYSASFYPCM+ILYECYEDV SGSEIR
Sbjct: 348 ECITGTISRTISTQGMLAVYNSLSEEGKKDFETAYSASFYPCMEILYECYEDVQSGSEIR 407
Query: 362 SVVLAGRRFYEKEGLPAFPMGKIDQTRMWKVGERVRSTRPAGDLGPLNPFTAGVFVALMM 541
SVVLAGRRFYEKEGLPAFPMG IDQTRMWKVGERVR +RPAGDLGPL PFTAGV+VALMM
Sbjct: 408 SVVLAGRRFYEKEGLPAFPMGNIDQTRMWKVGERVRKSRPAGDLGPLYPFTAGVYVALMM 467
Query: 542 AQIEVLRKKGHSYSEIINESVIESVDSLNPFMHARGVSFMVDNCSTTARLGSRKWAPRFD 721
AQIE+LRKKGHSYSEIINESVIESVDSLNPFMHARGVSFMVDNCSTTARLGSRKWAPRFD
Sbjct: 468 AQIEILRKKGHSYSEIINESVIESVDSLNPFMHARGVSFMVDNCSTTARLGSRKWAPRFD 527
Query: 722 YVLTQQALVAVDNAAPINQDLISNFLSDPVHGAIEVCAELRPTLDISVPPDADFVRPELR 901
Y+LTQQALVAVD+ A IN+DLISNF SDPVHGAIEVCA+LRPT+DISVP DADFVRPELR
Sbjct: 528 YILTQQALVAVDSGAAINRDLISNFFSDPVHGAIEVCAQLRPTVDISVPADADFVRPELR 587
Query: 902 QSSN 913
QSSN
Sbjct: 588 QSSN 591
>pir||S30145 ketol-acid reductoisomerase (EC 1.1.1.86) precursor - Arabidopsis
thaliana
Length = 591
Score = 560 bits (1444), Expect = e-158
Identities = 276/304 (90%), Positives = 291/304 (94%)
Frame = +2
Query: 2 WSVALGSPFTFATTLEQEYKSDIFGERGILLGAVHGIVESLFRRYTENGMAEDLAYKNTV 181
WSVALGSPFTFATTLEQEY+SDIFGERGILLGAVHGIVESLFRRYTENGM+EDLAYKNTV
Sbjct: 288 WSVALGSPFTFATTLEQEYRSDIFGERGILLGAVHGIVESLFRRYTENGMSEDLAYKNTV 347
Query: 182 ESITGIISKTISTKGMLAVYNSFSEEGKREFEKAYSASFYPCMDILYECYEDVASGSEIR 361
E ITG IS+TIST+GMLAVYNS SEEGK++FE AYSASFYPCM+ILYECYEDV SGSEIR
Sbjct: 348 ECITGTISRTISTQGMLAVYNSLSEEGKKDFETAYSASFYPCMEILYECYEDVQSGSEIR 407
Query: 362 SVVLAGRRFYEKEGLPAFPMGKIDQTRMWKVGERVRSTRPAGDLGPLNPFTAGVFVALMM 541
SVVLAGRRFYEKEGLPAFPMG IDQTRMWKVGERVR +RPAGDLGPL PFTAGV+VALMM
Sbjct: 408 SVVLAGRRFYEKEGLPAFPMGNIDQTRMWKVGERVRKSRPAGDLGPLYPFTAGVYVALMM 467
Query: 542 AQIEVLRKKGHSYSEIINESVIESVDSLNPFMHARGVSFMVDNCSTTARLGSRKWAPRFD 721
AQIE+LRKKGHSYSEIINESVIESVDSLNPFMHARGVSFMVDNCSTTARLGSRKWAPRFD
Sbjct: 468 AQIEILRKKGHSYSEIINESVIESVDSLNPFMHARGVSFMVDNCSTTARLGSRKWAPRFD 527
Query: 722 YVLTQQALVAVDNAAPINQDLISNFLSDPVHGAIEVCAELRPTLDISVPPDADFVRPELR 901
Y+LTQQALVAVD+ A IN+DLISNF SDPVHGAIEVCA+LRPT+DISVP D DFVRPELR
Sbjct: 528 YILTQQALVAVDSGAAINRDLISNFFSDPVHGAIEVCAQLRPTVDISVPADVDFVRPELR 587
Query: 902 QSSN 913
QSSN
Sbjct: 588 QSSN 591
>emb|CAA48253.1| ketol-acid reductoisomerase [Arabidopsis thaliana]
Length = 591
Score = 560 bits (1444), Expect = e-158
Identities = 276/304 (90%), Positives = 291/304 (94%)
Frame = +2
Query: 2 WSVALGSPFTFATTLEQEYKSDIFGERGILLGAVHGIVESLFRRYTENGMAEDLAYKNTV 181
WSVALGSPFTFATTLEQEY+SDIFGERGILLGAVHGIVESLFRRYTENGM+EDLAYKNTV
Sbjct: 288 WSVALGSPFTFATTLEQEYRSDIFGERGILLGAVHGIVESLFRRYTENGMSEDLAYKNTV 347
Query: 182 ESITGIISKTISTKGMLAVYNSFSEEGKREFEKAYSASFYPCMDILYECYEDVASGSEIR 361
E ITG IS+TIST+GMLAVYNS SEEGK++FE AYSASFYPCM+ILYECYEDV SGSEIR
Sbjct: 348 ECITGTISRTISTQGMLAVYNSLSEEGKKDFETAYSASFYPCMEILYECYEDVQSGSEIR 407
Query: 362 SVVLAGRRFYEKEGLPAFPMGKIDQTRMWKVGERVRSTRPAGDLGPLNPFTAGVFVALMM 541
SVVLAGRRFYEKEGLPAFPMG IDQTRMWKVGERVR +RPAGDLGPL PFTAGV+VALMM
Sbjct: 408 SVVLAGRRFYEKEGLPAFPMGNIDQTRMWKVGERVRKSRPAGDLGPLYPFTAGVYVALMM 467
Query: 542 AQIEVLRKKGHSYSEIINESVIESVDSLNPFMHARGVSFMVDNCSTTARLGSRKWAPRFD 721
AQIE+LRKKGHSYSEIINESVIESVDSLNPFMHARGVSFMVDNCSTTARLGSRKWAPRFD
Sbjct: 468 AQIEILRKKGHSYSEIINESVIESVDSLNPFMHARGVSFMVDNCSTTARLGSRKWAPRFD 527
Query: 722 YVLTQQALVAVDNAAPINQDLISNFLSDPVHGAIEVCAELRPTLDISVPPDADFVRPELR 901
Y+LTQQALVAVD+ A IN+DLISNF SDPVHGAIEVCA+LRPT+DISVP D DFVRPELR
Sbjct: 528 YILTQQALVAVDSGAAINRDLISNFFSDPVHGAIEVCAQLRPTVDISVPADVDFVRPELR 587
Query: 902 QSSN 913
QSSN
Sbjct: 588 QSSN 591
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,097,122,218
Number of Sequences: 1393205
Number of extensions: 24786670
Number of successful extensions: 59995
Number of sequences better than 10.0: 142
Number of HSP's better than 10.0 without gapping: 56375
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 59933
length of database: 448,689,247
effective HSP length: 126
effective length of database: 273,145,417
effective search space used: 81124188849
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)