Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
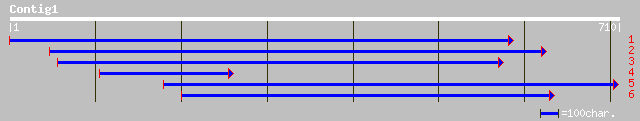
Query= KMC005692A_C01 KMC005692A_c01
(706 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAO37965.1| putative serine/threonine protein kinase [Oryza s... 228 6e-59
gb|AAM91528.1| putative casein kinase [Arabidopsis thaliana] 226 2e-58
gb|AAF00624.1|AC009540_1 unknown protein, 5' partial [Arabidopsi... 226 2e-58
ref|NP_187044.1| putative casein kinase; protein id: At3g03940.1... 226 2e-58
dbj|BAB56099.1| casein kinase-like protein [Oryza sativa (japoni... 226 3e-58
>gb|AAO37965.1| putative serine/threonine protein kinase [Oryza sativa (japonica
cultivar-group)]
Length = 707
Score = 228 bits (581), Expect = 6e-59
Identities = 104/126 (82%), Positives = 116/126 (91%)
Frame = -1
Query: 706 KVSDSFPYKWINRKWKEGFQVTSMATAGTRWGVVMSRNAGFSDQVVELDFLYPSEGIHKR 527
KVS+SFPYKWIN+KWKEGF VTSMATAG RWGVVMSRNAG+S QVVELDFLYPSEGIH+R
Sbjct: 582 KVSESFPYKWINKKWKEGFHVTSMATAGNRWGVVMSRNAGYSHQVVELDFLYPSEGIHRR 641
Query: 526 WDNGYRITATAATWDQSALILSIPRRKPNDETQETLRTSQFPSTHVKEKWAKNLYLSCLC 347
W+ GYRIT+TAAT DQ+A ILSIP+RKP DETQETLRTS FPS HVKEKW+KNLY++ +C
Sbjct: 642 WETGYRITSTAATPDQAAFILSIPKRKPMDETQETLRTSSFPSNHVKEKWSKNLYIASIC 701
Query: 346 YGRTVC 329
YGRTVC
Sbjct: 702 YGRTVC 707
Score = 54.3 bits (129), Expect = 2e-06
Identities = 30/120 (25%), Positives = 65/120 (54%), Gaps = 2/120 (1%)
Frame = -1
Query: 703 VSDSFPYKWINRKWKEGFQVTSMATAGTRWGVVMSRNAGFSDQVVELDFLY-PSEGIHKR 527
V+DS ++ I + ++G ++ ++++ W ++M GF QV EL ++ + I ++
Sbjct: 489 VADSRLHQHIEKGNEDGLYISCVSSSANFWALIMDAGTGFCSQVYELSQVFLHKDWIMEQ 548
Query: 526 WDNGYRITATAATWDQSALILSIPRRKPNDETQETLRTSQ-FPSTHVKEKWAKNLYLSCL 350
W+ Y ITA A + S+L++ K TQ++ + S+ FP + +KW + +++ +
Sbjct: 549 WEKNYYITAIAGATNGSSLVV---MSKGTPYTQQSYKVSESFPYKWINKKWKEGFHVTSM 605
>gb|AAM91528.1| putative casein kinase [Arabidopsis thaliana]
Length = 303
Score = 226 bits (577), Expect = 2e-58
Identities = 103/126 (81%), Positives = 118/126 (92%)
Frame = -1
Query: 706 KVSDSFPYKWINRKWKEGFQVTSMATAGTRWGVVMSRNAGFSDQVVELDFLYPSEGIHKR 527
KVSDSFP+KWIN+KWKEGF VTSM TAG+RWGVVMSRN+G+S+QVVELDFLYPSEGIH+R
Sbjct: 178 KVSDSFPFKWINKKWKEGFHVTSMTTAGSRWGVVMSRNSGYSEQVVELDFLYPSEGIHRR 237
Query: 526 WDNGYRITATAATWDQSALILSIPRRKPNDETQETLRTSQFPSTHVKEKWAKNLYLSCLC 347
W++GYRIT+ AAT DQ+ALILSIP+RK DETQETLRTS FPSTHVKEKWAKNLY++ +C
Sbjct: 238 WESGYRITSMAATADQAALILSIPKRKITDETQETLRTSAFPSTHVKEKWAKNLYIASIC 297
Query: 346 YGRTVC 329
YGRTVC
Sbjct: 298 YGRTVC 303
Score = 50.1 bits (118), Expect = 3e-05
Identities = 26/106 (24%), Positives = 57/106 (53%), Gaps = 2/106 (1%)
Frame = -1
Query: 661 KEGFQVTSMATAGTRWGVVMSRNAGFSDQVVELDFLY-PSEGIHKRWDNGYRITATAATW 485
++G ++ +A++ W ++M GFS QV EL ++ + I ++W+ Y I++ A
Sbjct: 99 EDGLFISCVASSANLWAIIMDAGTGFSSQVYELSSVFLHKDWIMEQWEKNYYISSIAGAN 158
Query: 484 DQSALILSIPRRKPNDETQETLRTS-QFPSTHVKEKWAKNLYLSCL 350
+ S+L++ K TQ++ + S FP + +KW + +++ +
Sbjct: 159 NGSSLVV---MAKGTPYTQQSYKVSDSFPFKWINKKWKEGFHVTSM 201
>gb|AAF00624.1|AC009540_1 unknown protein, 5' partial [Arabidopsis thaliana]
Length = 494
Score = 226 bits (577), Expect = 2e-58
Identities = 103/126 (81%), Positives = 118/126 (92%)
Frame = -1
Query: 706 KVSDSFPYKWINRKWKEGFQVTSMATAGTRWGVVMSRNAGFSDQVVELDFLYPSEGIHKR 527
KVSDSFP+KWIN+KWKEGF VTSM TAG+RWGVVMSRN+G+S+QVVELDFLYPSEGIH+R
Sbjct: 369 KVSDSFPFKWINKKWKEGFHVTSMTTAGSRWGVVMSRNSGYSEQVVELDFLYPSEGIHRR 428
Query: 526 WDNGYRITATAATWDQSALILSIPRRKPNDETQETLRTSQFPSTHVKEKWAKNLYLSCLC 347
W++GYRIT+ AAT DQ+ALILSIP+RK DETQETLRTS FPSTHVKEKWAKNLY++ +C
Sbjct: 429 WESGYRITSMAATADQAALILSIPKRKITDETQETLRTSAFPSTHVKEKWAKNLYIASIC 488
Query: 346 YGRTVC 329
YGRTVC
Sbjct: 489 YGRTVC 494
Score = 50.1 bits (118), Expect = 3e-05
Identities = 26/106 (24%), Positives = 57/106 (53%), Gaps = 2/106 (1%)
Frame = -1
Query: 661 KEGFQVTSMATAGTRWGVVMSRNAGFSDQVVELDFLY-PSEGIHKRWDNGYRITATAATW 485
++G ++ +A++ W ++M GFS QV EL ++ + I ++W+ Y I++ A
Sbjct: 290 EDGLFISCVASSANLWAIIMDAGTGFSSQVYELSSVFLHKDWIMEQWEKNYYISSIAGAN 349
Query: 484 DQSALILSIPRRKPNDETQETLRTS-QFPSTHVKEKWAKNLYLSCL 350
+ S+L++ K TQ++ + S FP + +KW + +++ +
Sbjct: 350 NGSSLVV---MAKGTPYTQQSYKVSDSFPFKWINKKWKEGFHVTSM 392
>ref|NP_187044.1| putative casein kinase; protein id: At3g03940.1 [Arabidopsis
thaliana] gi|6223639|gb|AAF05853.1|AC011698_4 putative
casein kinase [Arabidopsis thaliana]
Length = 701
Score = 226 bits (577), Expect = 2e-58
Identities = 103/126 (81%), Positives = 118/126 (92%)
Frame = -1
Query: 706 KVSDSFPYKWINRKWKEGFQVTSMATAGTRWGVVMSRNAGFSDQVVELDFLYPSEGIHKR 527
KVSDSFP+KWIN+KWKEGF VTSM TAG+RWGVVMSRN+G+S+QVVELDFLYPSEGIH+R
Sbjct: 576 KVSDSFPFKWINKKWKEGFHVTSMTTAGSRWGVVMSRNSGYSEQVVELDFLYPSEGIHRR 635
Query: 526 WDNGYRITATAATWDQSALILSIPRRKPNDETQETLRTSQFPSTHVKEKWAKNLYLSCLC 347
W++GYRIT+ AAT DQ+ALILSIP+RK DETQETLRTS FPSTHVKEKWAKNLY++ +C
Sbjct: 636 WESGYRITSMAATADQAALILSIPKRKITDETQETLRTSAFPSTHVKEKWAKNLYIASIC 695
Query: 346 YGRTVC 329
YGRTVC
Sbjct: 696 YGRTVC 701
Score = 50.1 bits (118), Expect = 3e-05
Identities = 26/106 (24%), Positives = 57/106 (53%), Gaps = 2/106 (1%)
Frame = -1
Query: 661 KEGFQVTSMATAGTRWGVVMSRNAGFSDQVVELDFLY-PSEGIHKRWDNGYRITATAATW 485
++G ++ +A++ W ++M GFS QV EL ++ + I ++W+ Y I++ A
Sbjct: 497 EDGLFISCVASSANLWAIIMDAGTGFSSQVYELSSVFLHKDWIMEQWEKNYYISSIAGAN 556
Query: 484 DQSALILSIPRRKPNDETQETLRTS-QFPSTHVKEKWAKNLYLSCL 350
+ S+L++ K TQ++ + S FP + +KW + +++ +
Sbjct: 557 NGSSLVV---MAKGTPYTQQSYKVSDSFPFKWINKKWKEGFHVTSM 599
>dbj|BAB56099.1| casein kinase-like protein [Oryza sativa (japonica cultivar-group)]
gi|20160458|dbj|BAB89411.1| casein kinase-like protein
[Oryza sativa (japonica cultivar-group)]
Length = 621
Score = 226 bits (575), Expect = 3e-58
Identities = 105/125 (84%), Positives = 115/125 (92%)
Frame = -1
Query: 706 KVSDSFPYKWINRKWKEGFQVTSMATAGTRWGVVMSRNAGFSDQVVELDFLYPSEGIHKR 527
KVSDSFP+KWIN+KWKEGF VT++ATAG+RW VVMSRNAGF+ QVVELDFLYPSEGIH+R
Sbjct: 496 KVSDSFPFKWINKKWKEGFYVTALATAGSRWAVVMSRNAGFTHQVVELDFLYPSEGIHQR 555
Query: 526 WDNGYRITATAATWDQSALILSIPRRKPNDETQETLRTSQFPSTHVKEKWAKNLYLSCLC 347
WD+GYRITATAAT DQ ALILSIPRRKPNDETQETLRTS FP HVKEKWAKNLYL +C
Sbjct: 556 WDSGYRITATAATCDQVALILSIPRRKPNDETQETLRTSAFPGQHVKEKWAKNLYLGSIC 615
Query: 346 YGRTV 332
YGR+V
Sbjct: 616 YGRSV 620
Score = 42.4 bits (98), Expect = 0.006
Identities = 24/91 (26%), Positives = 46/91 (50%), Gaps = 2/91 (2%)
Frame = -1
Query: 616 WGVVMSRNAGFSDQVVELD-FLYPSEGIHKRWDNGYRITATAATWDQSALILSIPRRKPN 440
W ++M GF+ QV EL + E I ++W+ Y IT+ A + + S++++
Sbjct: 432 WALIMDAGTGFTAQVHELSHYFLHKEWIMEQWERNYYITSLAGSNNGSSVVI---MSTGT 488
Query: 439 DETQETLRTS-QFPSTHVKEKWAKNLYLSCL 350
Q++ + S FP + +KW + Y++ L
Sbjct: 489 PYAQQSYKVSDSFPFKWINKKWKEGFYVTAL 519
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 640,409,529
Number of Sequences: 1393205
Number of extensions: 14658275
Number of successful extensions: 35558
Number of sequences better than 10.0: 24
Number of HSP's better than 10.0 without gapping: 34269
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 35513
length of database: 448,689,247
effective HSP length: 120
effective length of database: 281,504,647
effective search space used: 32091529758
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)