Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
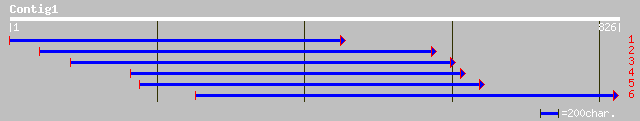
Query= KMC005686A_C01 KMC005686A_c01
(823 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
sp|Q43075|SPE1_PEA ARGININE DECARBOXYLASE (ARGDC) (ADC) gi|21298... 218 1e-55
emb|CAA65585.1| arginine decarboxylase [Vitis vinifera] 193 3e-48
gb|AAF42970.1|AF127239_1 arginine decarboxylase 1 [Nicotiana tab... 191 1e-47
emb|CAB64599.1| arginine decarboxylase 1 [Datura stramonium] 189 6e-47
gb|AAF42972.1|AF127241_1 arginine decarboxylase 2 [Nicotiana tab... 184 1e-45
>sp|Q43075|SPE1_PEA ARGININE DECARBOXYLASE (ARGDC) (ADC) gi|2129876|pir||S59553 arginine
decarboxylase (EC 4.1.1.19) - garden pea
gi|609220|emb|CAA85773.1| arginine decarboxylase [Pisum
sativum]
Length = 728
Score = 218 bits (554), Expect = 1e-55
Identities = 110/160 (68%), Positives = 126/160 (78%), Gaps = 8/160 (5%)
Frame = -3
Query: 821 LHELED--GHYYLGMFLGGAYEEALGGVHNLFGGPSVVRVVQSDGPHGFAVTRAVAGPSC 648
LHE+E G YYLGMFLGG+YEEALGG+HNLFGGPSVVRV+QSDGPHGFAVTRAVAG SC
Sbjct: 570 LHEMEGHGGGYYLGMFLGGSYEEALGGLHNLFGGPSVVRVLQSDGPHGFAVTRAVAGSSC 629
Query: 647 GDVLRAMQHQPELMFETLKHRVLEF-----DQDEVNAAAVVASLARSFDKMPYLVAAKPC 483
DVLR MQH+P+LMFETLKHR LEF D VNA + SLA+SFD MPYLV++ C
Sbjct: 630 ADVLRVMQHEPQLMFETLKHRALEFCGQHDDDSVVNAGVLANSLAQSFDNMPYLVSSTTC 689
Query: 482 CFNAV-SNGGFYHCSGDDFSADSGGGGGGSAGGEEEHWSY 366
C NA+ +N GFY+CSGDDFSAD+ S GE+E+WSY
Sbjct: 690 CLNALTNNNGFYYCSGDDFSADT-VSVATSVAGEDENWSY 728
>emb|CAA65585.1| arginine decarboxylase [Vitis vinifera]
Length = 630
Score = 193 bits (490), Expect = 3e-48
Identities = 99/164 (60%), Positives = 119/164 (72%), Gaps = 9/164 (5%)
Frame = -3
Query: 821 LHELE--------DGHYYLGMFLGGAYEEALGGVHNLFGGPSVVRVVQSDGPHGFAVTRA 666
LHELE G YYLGMFLGGAYEEALGG+HNLFGGPSVVRV+QSDGPH FAVTRA
Sbjct: 475 LHELEGSDVVFGGSGKYYLGMFLGGAYEEALGGLHNLFGGPSVVRVLQSDGPHSFAVTRA 534
Query: 665 VAGPSCGDVLRAMQHQPELMFETLKHRVLEF-DQDEVNAAAVVASLARSFDKMPYLVAAK 489
+ GPSCGDVLR MQH+PELMFETLKHR E +D + ++ + LA SF KMPYLVA
Sbjct: 535 MPGPSCGDVLRVMQHEPELMFETLKHRAEECGHEDGMTNGSLASGLALSFHKMPYLVAGS 594
Query: 488 PCCFNAVSNGGFYHCSGDDFSADSGGGGGGSAGGEEEHWSYCCA 357
CC ++N G+Y+ + D++S + SA G+++HWSYC A
Sbjct: 595 SCC---MTNSGYYYGNEDNYSRVA-----DSAAGDDDHWSYCFA 630
>gb|AAF42970.1|AF127239_1 arginine decarboxylase 1 [Nicotiana tabacum]
gi|7230373|gb|AAF42971.1|AF127240_1 arginine
decarboxylase 1 [Nicotiana tabacum]
Length = 720
Score = 191 bits (484), Expect = 1e-47
Identities = 102/165 (61%), Positives = 120/165 (71%), Gaps = 10/165 (6%)
Frame = -3
Query: 821 LHEL----EDGHYYLGMFLGGAYEEALGGVHNLFGGPSVVRVVQSDGPHGFAVTRAVAGP 654
LHEL + G YYLGMFLGGAYEEALGG+HNLFGGPSVVRVVQSD H FA++R+V GP
Sbjct: 563 LHELGSNGDGGGYYLGMFLGGAYEEALGGLHNLFGGPSVVRVVQSDSAHSFAMSRSVPGP 622
Query: 653 SCGDVLRAMQHQPELMFETLKHRVLEFDQDE----VNAAAVVASLARSFDKMPYLVAAKP 486
SC DVLRAMQH+PELMFETLKHR EF + E + A++ +SLA+SF MPYLVA
Sbjct: 623 SCADVLRAMQHEPELMFETLKHRAEEFLEQEEDKGLAIASLASSLAQSFHNMPYLVAPAS 682
Query: 485 CCFNAV--SNGGFYHCSGDDFSADSGGGGGGSAGGEEEHWSYCCA 357
CCF AV +NGG+ + D+ +AD SA GE+E WSYC A
Sbjct: 683 CCFTAVTANNGGYNYYYSDENAAD-------SATGEDEIWSYCTA 720
>emb|CAB64599.1| arginine decarboxylase 1 [Datura stramonium]
Length = 724
Score = 189 bits (479), Expect = 6e-47
Identities = 101/166 (60%), Positives = 121/166 (72%), Gaps = 11/166 (6%)
Frame = -3
Query: 821 LHEL--EDGHYYLGMFLGGAYEEALGGVHNLFGGPSVVRVVQSDGPHGFAVTRAVAGPSC 648
LHEL + G YYLGMFLGGAYEEALGG+HNLFGGPSVVRV+QSD PH FAVTR+V GPSC
Sbjct: 567 LHELGSDGGRYYLGMFLGGAYEEALGGLHNLFGGPSVVRVLQSDSPHSFAVTRSVPGPSC 626
Query: 647 GDVLRAMQHQPELMFETLKHRVLEFDQDE-------VNAAAVVASLARSFDKMPYLVAAK 489
DVLRAMQ +PELMFETLKHR E+ + E ++ A++ +SLA+SF MPYLVA
Sbjct: 627 ADVLRAMQFEPELMFETLKHRAEEYLEQEEKEEDKSMSFASLTSSLAQSFHNMPYLVAPS 686
Query: 488 PCCFNAV--SNGGFYHCSGDDFSADSGGGGGGSAGGEEEHWSYCCA 357
CCF A +NGG+Y+ S +D +AD A GE++ WSYC A
Sbjct: 687 SCCFTAATGNNGGYYYYS-EDKAAD-------CATGEDDIWSYCTA 724
>gb|AAF42972.1|AF127241_1 arginine decarboxylase 2 [Nicotiana tabacum]
Length = 721
Score = 184 bits (467), Expect = 1e-45
Identities = 101/165 (61%), Positives = 118/165 (71%), Gaps = 10/165 (6%)
Frame = -3
Query: 821 LHEL----EDGHYYLGMFLGGAYEEALGGVHNLFGGPSVVRVVQSDGPHGFAVTRAVAGP 654
LHEL + G YYLGMFLGGAYEEALGG+HNLFGGPSVVRVVQSD H FA+TR+V GP
Sbjct: 564 LHELGSNGDGGGYYLGMFLGGAYEEALGGLHNLFGGPSVVRVVQSDSAHSFAMTRSVPGP 623
Query: 653 SCGDVLRAMQHQPELMFETLKHRVLEF-DQDEVNAAAV---VASLARSFDKMPYLVAAKP 486
SC DVLRAMQH+PELMFETLKHR EF +Q++ AV +S+A+SF MPYLVA
Sbjct: 624 SCADVLRAMQHEPELMFETLKHRAEEFLEQEDDKGLAVESLASSVAQSFHNMPYLVAPSS 683
Query: 485 CCFNAV--SNGGFYHCSGDDFSADSGGGGGGSAGGEEEHWSYCCA 357
C F A +NGG+ + D+ +AD SA GE+E WSYC A
Sbjct: 684 CRFTAATDNNGGYNYYYSDENAAD-------SATGEDEIWSYCTA 721
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 736,022,050
Number of Sequences: 1393205
Number of extensions: 18019682
Number of successful extensions: 165905
Number of sequences better than 10.0: 301
Number of HSP's better than 10.0 without gapping: 82849
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 138199
length of database: 448,689,247
effective HSP length: 121
effective length of database: 280,111,442
effective search space used: 42576939184
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)