Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC005674A_C01 KMC005674A_c01
(805 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
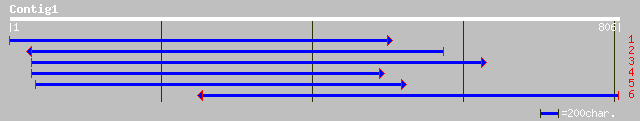
ref|NP_173653.1| UDP-glucose glucosyltransferase; protein id: At... 263 2e-69
gb|AAL32657.1| UDP-glucose glucosyltransferase [Arabidopsis thal... 261 6e-69
pir||F86356 T16E15.2 protein - Arabidopsis thaliana gi|9392678|g... 252 5e-66
dbj|BAB86928.1| glucosyltransferase-10 [Vigna angularis] 249 3e-65
ref|NP_173652.1| UDP-glucose glucosyltransferase, putative; prot... 248 3e-65
>ref|NP_173653.1| UDP-glucose glucosyltransferase; protein id: At1g22360.1, supported
by cDNA: gi_12083243, supported by cDNA: gi_17065005,
supported by cDNA: gi_20260017, supported by cDNA:
gi_3928542 [Arabidopsis thaliana]
gi|25286823|pir||E86356 hypothetical protein T16E15.3 -
Arabidopsis thaliana gi|3928543|dbj|BAA34687.1|
UDP-glucose glucosyltransferase [Arabidopsis thaliana]
gi|9392679|gb|AAF87256.1|AC068562_3 Identical to
UDP-glucose glucosyltransferase from Arabidopsis
thaliana gb|AB016819 and contains a UDP-glucosyl
transferase PF|00201 domain. ESTs gb|T46254, gb|R83990,
gb|H37246, gb|W43072, gb|R90721, gb|R90712, gb|AA712612,
gb|AA404770 come from this gene
gi|12083244|gb|AAG48781.1|AF332418_1 putative
UDP-glucose glucosyltransferase [Arabidopsis thaliana]
Length = 481
Score = 263 bits (673), Expect = 2e-69
Identities = 120/213 (56%), Positives = 152/213 (71%)
Frame = +2
Query: 68 SSKPHAVCVPYPAQGHVNPFMQLTKLLRCMGFHITFVNTEYNHKRLAKSLGPEFVKGLPD 247
+ K H VCVPYPAQGH+NP M++ KLL GFHITFVNT YNH RL +S GP V GLP
Sbjct: 6 AQKQHVVCVPYPAQGHINPMMKVAKLLYAKGFHITFVNTVYNHNRLLRSRGPNAVDGLPS 65
Query: 248 FRFETIPDGLPPPEKDATQSIPLLSQSTSKHCYEPFKELVNKLNSSPEGPPVSCVITDGV 427
FRFE+IPDGLP + D TQ IP L +ST KHC PFKEL+ ++N+ + PPVSC+++DG
Sbjct: 66 FRFESIPDGLPETDVDVTQDIPTLCESTMKHCLAPFKELLRQINARDDVPPVSCIVSDGC 125
Query: 428 MGFASSVANDLGIQEIQFWTASACGLLAYLQFEELLKRDIVPLKDENFMSDGTLDTTIDW 607
M F A +LG+ E+ FWT SACG LAYL + +++ + P+KDE++++ LDT IDW
Sbjct: 126 MSFTLDAAEELGVPEVLFWTTSACGFLAYLYYYRFIEKGLSPIKDESYLTKEHLDTKIDW 185
Query: 608 ISGMKNMRLKDFPSFVRVTDLKDDIMYKFFISE 706
I MKN+RLKD PSF+R T+ DDIM F I E
Sbjct: 186 IPSMKNLRLKDIPSFIRTTN-PDDIMLNFIIRE 217
>gb|AAL32657.1| UDP-glucose glucosyltransferase [Arabidopsis thaliana]
gi|20260018|gb|AAM13356.1| UDP-glucose
glucosyltransferase [Arabidopsis thaliana]
Length = 481
Score = 261 bits (668), Expect = 6e-69
Identities = 119/213 (55%), Positives = 152/213 (70%)
Frame = +2
Query: 68 SSKPHAVCVPYPAQGHVNPFMQLTKLLRCMGFHITFVNTEYNHKRLAKSLGPEFVKGLPD 247
+ K H VCVPYPAQGH+NP M++ KLL GFHITFVNT YNH RL +S GP V GLP
Sbjct: 6 AQKQHVVCVPYPAQGHINPMMKVAKLLYAKGFHITFVNTVYNHNRLLRSRGPNAVDGLPS 65
Query: 248 FRFETIPDGLPPPEKDATQSIPLLSQSTSKHCYEPFKELVNKLNSSPEGPPVSCVITDGV 427
FRFE+IPDGLP + D TQ IP L +ST KHC PFKEL+ ++N+ + PPVSC+++DG
Sbjct: 66 FRFESIPDGLPETDVDVTQDIPTLCESTMKHCLAPFKELLRQINARDDVPPVSCIVSDGC 125
Query: 428 MGFASSVANDLGIQEIQFWTASACGLLAYLQFEELLKRDIVPLKDENFMSDGTLDTTIDW 607
M F A +LG+ E+ FWT SACG LAYL + +++ + P+KDE++++ LDT IDW
Sbjct: 126 MSFTLDAAEELGVPEVLFWTTSACGFLAYLYYYRFIEKGLSPIKDESYLTKEHLDTKIDW 185
Query: 608 ISGMKNMRLKDFPSFVRVTDLKDDIMYKFFISE 706
I MK++RLKD PSF+R T+ DDIM F I E
Sbjct: 186 IPSMKDLRLKDIPSFIRTTN-PDDIMLNFIIRE 217
>pir||F86356 T16E15.2 protein - Arabidopsis thaliana
gi|9392678|gb|AAF87255.1|AC068562_2 Strong similarity to
UDP-glucose glucosyltransferase from Arabidopsis
thaliana gb|AB016819 and contains a UDP-glucosyl
transferase PF|00201 domain. ESTs gb|U74128,
gb|AA713257 come from this gene
Length = 479
Score = 252 bits (643), Expect = 5e-66
Identities = 112/214 (52%), Positives = 152/214 (70%)
Frame = +2
Query: 65 ASSKPHAVCVPYPAQGHVNPFMQLTKLLRCMGFHITFVNTEYNHKRLAKSLGPEFVKGLP 244
+ KPH VC+P+PAQGH+NP +++ KLL GFH+TFVNT YNH RL +S GP + GLP
Sbjct: 8 SGQKPHVVCIPFPAQGHINPMLKVAKLLYARGFHVTFVNTNYNHNRLIRSRGPNSLDGLP 67
Query: 245 DFRFETIPDGLPPPEKDATQSIPLLSQSTSKHCYEPFKELVNKLNSSPEGPPVSCVITDG 424
FRFE+IPDGLP KD Q +P L +ST K+C PFKEL+ ++N++ + PPVSC+++DG
Sbjct: 68 SFRFESIPDGLPEENKDVMQDVPTLCESTMKNCLAPFKELLRRINTTKDVPPVSCIVSDG 127
Query: 425 VMGFASSVANDLGIQEIQFWTASACGLLAYLQFEELLKRDIVPLKDENFMSDGTLDTTID 604
VM F A +LG+ ++ FWT SACG LAYL F +++ + P+KDE+ +LDT I+
Sbjct: 128 VMSFTLDAAEELGVPDVLFWTPSACGFLAYLHFYRFIEKGLSPIKDES-----SLDTKIN 182
Query: 605 WISGMKNMRLKDFPSFVRVTDLKDDIMYKFFISE 706
WI MKN+ LKD PSF+R T+ +DIM FF+ E
Sbjct: 183 WIPSMKNLGLKDIPSFIRATN-TEDIMLNFFVHE 215
>dbj|BAB86928.1| glucosyltransferase-10 [Vigna angularis]
Length = 485
Score = 249 bits (636), Expect = 3e-65
Identities = 115/210 (54%), Positives = 150/210 (70%)
Frame = +2
Query: 65 ASSKPHAVCVPYPAQGHVNPFMQLTKLLRCMGFHITFVNTEYNHKRLAKSLGPEFVKGLP 244
A K HAVCVP+PAQGH+NP ++L KLL GF+ITFVNT YNHKRL KS G + GLP
Sbjct: 6 AKEKQHAVCVPFPAQGHINPMLKLAKLLHFNGFYITFVNTHYNHKRLLKSRGLNSLNGLP 65
Query: 245 DFRFETIPDGLPPPEKDATQSIPLLSQSTSKHCYEPFKELVNKLNSSPEGPPVSCVITDG 424
FRFETIPDGLP PE + T +P L STS C F+ L++KLN+ P VSC+I+DG
Sbjct: 66 SFRFETIPDGLPEPEVEGTHHVPSLCDSTSTTCLPHFRNLLSKLNNESGVPAVSCIISDG 125
Query: 425 VMGFASSVANDLGIQEIQFWTASACGLLAYLQFEELLKRDIVPLKDENFMSDGTLDTTID 604
VM F + +LG+ + FWT+SACG + Y+ + +L++R IVP KD + +++G LDTTID
Sbjct: 126 VMSFTLDASQELGLPNVLFWTSSACGFMCYVHYHQLIQRGIVPFKDASDLTNGYLDTTID 185
Query: 605 WISGMKNMRLKDFPSFVRVTDLKDDIMYKF 694
W++G+K +RLKD PSF+R TD +DIM F
Sbjct: 186 WVAGIKEIRLKDIPSFIRTTD-PEDIMLNF 214
>ref|NP_173652.1| UDP-glucose glucosyltransferase, putative; protein id: At1g22340.1
[Arabidopsis thaliana] gi|25286820|pir||C86356
UDP-glucose glucosyltransferase homolog - Arabidopsis
thaliana gi|9392680|gb|AAF87257.1|AC068562_4 Strong
similarity to UDP-glucose glucosyltransferase from
Arabidopsis thaliana gb|AB016819 and contains a
UDP-glucosyl transferase PF|00201 domain
Length = 487
Score = 248 bits (632), Expect(2) = 3e-65
Identities = 111/215 (51%), Positives = 147/215 (67%)
Frame = +2
Query: 68 SSKPHAVCVPYPAQGHVNPFMQLTKLLRCMGFHITFVNTEYNHKRLAKSLGPEFVKGLPD 247
+ KPH VCVPYPAQGH+NP +++ KLL GFH+TFVNT YNH RL +S GP + G P
Sbjct: 9 AQKPHVVCVPYPAQGHINPMLKVAKLLYAKGFHVTFVNTLYNHNRLLRSRGPNALDGFPS 68
Query: 248 FRFETIPDGLPPPEKDATQSIPLLSQSTSKHCYEPFKELVNKLNSSPEGPPVSCVITDGV 427
FRFE+IPDGLP + D TQ P + S K+C PFKE++ ++N + PPVSC+++DGV
Sbjct: 69 FRFESIPDGLPETDGDRTQHTPTVCMSIEKNCLAPFKEILRRINDKDDVPPVSCIVSDGV 128
Query: 428 MGFASSVANDLGIQEIQFWTASACGLLAYLQFEELLKRDIVPLKDENFMSDGTLDTTIDW 607
M F A +LG+ E+ FWT SACG + L F +++ + P KDE++MS LDT IDW
Sbjct: 129 MSFTLDAAEELGVPEVIFWTNSACGFMTILHFYLFIEKGLSPFKDESYMSKEHLDTVIDW 188
Query: 608 ISGMKNMRLKDFPSFVRVTDLKDDIMYKFFISETQ 712
I MKN+RLKD PS++R T+ D+IM F I E +
Sbjct: 189 IPSMKNLRLKDIPSYIRTTN-PDNIMLNFLIREVE 222
Score = 23.5 bits (49), Expect(2) = 3e-65
Identities = 8/20 (40%), Positives = 16/20 (80%)
Frame = +3
Query: 732 IIINTYEELESEALDTLRVI 791
II+NT++ELE + + +++ I
Sbjct: 230 IILNTFDELEHDVIQSMQSI 249
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 724,874,742
Number of Sequences: 1393205
Number of extensions: 16013131
Number of successful extensions: 41393
Number of sequences better than 10.0: 266
Number of HSP's better than 10.0 without gapping: 39521
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 41271
length of database: 448,689,247
effective HSP length: 121
effective length of database: 280,111,442
effective search space used: 40896270532
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)