Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC005629A_C01 KMC005629A_c01
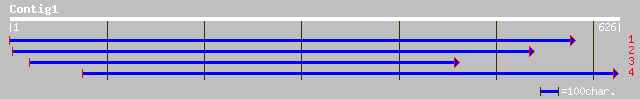
(623 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
pir||T12198 sucrose transport protein - fava bean gi|1935019|emb... 190 1e-47
gb|AAD41024.1| sucrose transport protein SUT1 [Pisum sativum] 188 4e-47
pir||S48789 sucrose transport protein - common tobacco gi|575351... 186 3e-46
pir||S43142 sucrose transport protein - castor bean gi|468562|em... 184 6e-46
emb|CAC19689.1| sucrose/proton symporter [Daucus carota] 183 2e-45
>pir||T12198 sucrose transport protein - fava bean gi|1935019|emb|CAB07811.1|
sucrose transport protein [Vicia faba]
Length = 523
Score = 190 bits (483), Expect = 1e-47
Identities = 92/148 (62%), Positives = 119/148 (80%), Gaps = 4/148 (2%)
Frame = -2
Query: 622 AVCMAMTMLITKVAEHDRRVSGGA-TIGRPTP---GVKAGALMFFAVLGIPLAITYSVPF 455
A+C+ +T+L+TK+A+H R+ + G +G P P G+KAGAL F+VLG+PLAITYS+PF
Sbjct: 376 AICLGLTVLVTKLAQHSRQYAPGTGALGDPLPPSEGIKAGALTLFSVLGVPLAITYSIPF 435
Query: 454 ALASIYSSSTGAGQGLSLGVLNVAIVIPQMIVSALNGPWDSLFGGGNLPAFMVGAVMAAV 275
ALASI+SS++GAGQGLSLGVLN+AIVIPQM VS L+GPWD+LFGGGNLPAF+VGAV A
Sbjct: 436 ALASIFSSTSGAGQGLSLGVLNLAIVIPQMFVSVLSGPWDALFGGGNLPAFVVGAVAALA 495
Query: 274 SAVLAMVLLPSPKPEEMAKASISASGFH 191
S +L+++LLPSP P+ S + GFH
Sbjct: 496 SGILSIILLPSPPPDMAKSVSATGGGFH 523
>gb|AAD41024.1| sucrose transport protein SUT1 [Pisum sativum]
Length = 524
Score = 188 bits (478), Expect = 4e-47
Identities = 93/148 (62%), Positives = 118/148 (78%), Gaps = 4/148 (2%)
Frame = -2
Query: 622 AVCMAMTMLITKVAEHDRRVSGGAT-IGRPTP---GVKAGALMFFAVLGIPLAITYSVPF 455
A+C+ +T+L+TK+A+H R+ + G + P P G+KAGAL F+VLGIPLAITYS+PF
Sbjct: 377 AICLGLTVLVTKLAQHSRQYAPGTGGLQDPLPPSGGIKAGALTLFSVLGIPLAITYSIPF 436
Query: 454 ALASIYSSSTGAGQGLSLGVLNVAIVIPQMIVSALNGPWDSLFGGGNLPAFMVGAVMAAV 275
ALASI+SS++GAGQGLSLGVLN+AIVIPQM VS L+GPWD+LFGGGNLPAF+VGAV A
Sbjct: 437 ALASIFSSTSGAGQGLSLGVLNLAIVIPQMFVSVLSGPWDALFGGGNLPAFVVGAVAALA 496
Query: 274 SAVLAMVLLPSPKPEEMAKASISASGFH 191
S +L+M+LLPSP P+ S + GFH
Sbjct: 497 SGILSMILLPSPPPDMAKSVSATGGGFH 524
>pir||S48789 sucrose transport protein - common tobacco
gi|575351|emb|CAA57727.1| sucrose transporter [Nicotiana
tabacum]
Length = 507
Score = 186 bits (471), Expect = 3e-46
Identities = 92/144 (63%), Positives = 113/144 (77%)
Frame = -2
Query: 622 AVCMAMTMLITKVAEHDRRVSGGATIGRPTPGVKAGALMFFAVLGIPLAITYSVPFALAS 443
AVCMA+T+L+TK+AE R+ T+ PT GVK GAL FAVLGIPLA+T+SVPFALAS
Sbjct: 366 AVCMALTVLVTKMAEKSRQYDAHGTLMAPTSGVKIGALTLFAVLGIPLAVTFSVPFALAS 425
Query: 442 IYSSSTGAGQGLSLGVLNVAIVIPQMIVSALNGPWDSLFGGGNLPAFMVGAVMAAVSAVL 263
I+SS+ G+GQGLSLGVLN+AIV+PQM+VS GPWD LFGGGNLP F+VGAV AA S +L
Sbjct: 426 IFSSNAGSGQGLSLGVLNLAIVVPQMLVSIAGGPWDDLFGGGNLPGFIVGAVAAAASGIL 485
Query: 262 AMVLLPSPKPEEMAKASISASGFH 191
A+ +LPSP + AK + + GFH
Sbjct: 486 ALTMLPSPPAD--AKPATTMGGFH 507
>pir||S43142 sucrose transport protein - castor bean gi|468562|emb|CAA83436.1|
sucrose carrier [Ricinus communis]
Length = 533
Score = 184 bits (468), Expect = 6e-46
Identities = 96/148 (64%), Positives = 117/148 (78%), Gaps = 4/148 (2%)
Frame = -2
Query: 622 AVCMAMTMLITKVAEHDRR---VSGGATIGRPTP-GVKAGALMFFAVLGIPLAITYSVPF 455
AVC+AMT+L+TK AE RR VSGGA + P P GVKAGAL FAV+G+P AITYS+PF
Sbjct: 387 AVCLAMTVLVTKQAESTRRFATVSGGAKVPLPPPSGVKAGALALFAVMGVPQAITYSIPF 446
Query: 454 ALASIYSSSTGAGQGLSLGVLNVAIVIPQMIVSALNGPWDSLFGGGNLPAFMVGAVMAAV 275
ALASI+S+++GAGQGLSLGVLN++IVIPQMIVS GPWD+LFGGGNLPAF+VGAV A
Sbjct: 447 ALASIFSNTSGAGQGLSLGVLNLSIVIPQMIVSVAAGPWDALFGGGNLPAFVVGAVAALA 506
Query: 274 SAVLAMVLLPSPKPEEMAKASISASGFH 191
S + A+ +LPSP+P +M A + FH
Sbjct: 507 SGIFALTMLPSPQP-DMPSAKALTAAFH 533
>emb|CAC19689.1| sucrose/proton symporter [Daucus carota]
Length = 515
Score = 183 bits (464), Expect = 2e-45
Identities = 90/141 (63%), Positives = 115/141 (80%)
Frame = -2
Query: 622 AVCMAMTMLITKVAEHDRRVSGGATIGRPTPGVKAGALMFFAVLGIPLAITYSVPFALAS 443
A+ + MT++++KVA+H R S + P+ GVKAGAL F++LGIPL+ITYS+PFALAS
Sbjct: 373 AIGLVMTVVVSKVAQHQREHSANGQLLPPSAGVKAGALSLFSILGIPLSITYSIPFALAS 432
Query: 442 IYSSSTGAGQGLSLGVLNVAIVIPQMIVSALNGPWDSLFGGGNLPAFMVGAVMAAVSAVL 263
IYSS +GAGQGLSLGVLN+AIV+PQMIVS L GP+DSLFGGGNLPAF+VGA+ AA+S VL
Sbjct: 433 IYSSGSGAGQGLSLGVLNLAIVVPQMIVSVLAGPFDSLFGGGNLPAFVVGAISAAISGVL 492
Query: 262 AMVLLPSPKPEEMAKASISAS 200
A+VLLP P + +K S+S +
Sbjct: 493 AIVLLPKPSKDAASKLSLSGT 513
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 592,581,870
Number of Sequences: 1393205
Number of extensions: 14758396
Number of successful extensions: 47117
Number of sequences better than 10.0: 117
Number of HSP's better than 10.0 without gapping: 43588
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 46942
length of database: 448,689,247
effective HSP length: 118
effective length of database: 284,291,057
effective search space used: 25301904073
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)