Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC005621A_C01 KMC005621A_c01
(565 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
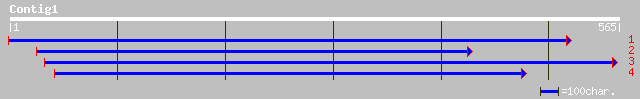
ref|NP_566534.1| kinesin-like protein TBK5, putative; protein id... 143 2e-33
dbj|BAB02671.1| contains similarity to kinesin heavy chain~gene_... 140 1e-32
dbj|BAB40710.1| BY-2 kinesin-like protein 10 [Nicotiana tabacum] 135 5e-31
dbj|BAB19066.1| kinesin-like protein [Oryza sativa (japonica cul... 116 2e-25
gb|AAK70904.1|AC087551_3 kinesin-like protein [Oryza sativa] 96 2e-19
>ref|NP_566534.1| kinesin-like protein TBK5, putative; protein id: At3g16060.1,
supported by cDNA: gi_15450500, supported by cDNA:
gi_16974324 [Arabidopsis thaliana]
gi|15450501|gb|AAK96543.1| AT3g16060/MSL1_10
[Arabidopsis thaliana] gi|16974325|gb|AAL31147.1|
AT3g16060/MSL1_10 [Arabidopsis thaliana]
Length = 684
Score = 143 bits (360), Expect = 2e-33
Identities = 70/80 (87%), Positives = 76/80 (94%)
Frame = -1
Query: 565 EEDLVNAHRTQVEETMNIVREEMNLLVEADQPGNQLDGYVTRLNAILSQKAAGILQLQTR 386
EEDLVNAHR QVE+TMNIV+EEMNLLVEADQPGNQLDGY++RLN ILSQKAAGILQLQ R
Sbjct: 605 EEDLVNAHRKQVEDTMNIVKEEMNLLVEADQPGNQLDGYISRLNTILSQKAAGILQLQNR 664
Query: 385 LAHFQKRLKEHNVLVSSHGY 326
LAHFQKRL+EHNVLVS+ GY
Sbjct: 665 LAHFQKRLREHNVLVSTTGY 684
>dbj|BAB02671.1| contains similarity to kinesin heavy chain~gene_id:MSL1.9
[Arabidopsis thaliana]
Length = 706
Score = 140 bits (353), Expect = 1e-32
Identities = 69/79 (87%), Positives = 75/79 (94%)
Frame = -1
Query: 565 EEDLVNAHRTQVEETMNIVREEMNLLVEADQPGNQLDGYVTRLNAILSQKAAGILQLQTR 386
EEDLVNAHR QVE+TMNIV+EEMNLLVEADQPGNQLDGY++RLN ILSQKAAGILQLQ R
Sbjct: 605 EEDLVNAHRKQVEDTMNIVKEEMNLLVEADQPGNQLDGYISRLNTILSQKAAGILQLQNR 664
Query: 385 LAHFQKRLKEHNVLVSSHG 329
LAHFQKRL+EHNVLVS+ G
Sbjct: 665 LAHFQKRLREHNVLVSTTG 683
>dbj|BAB40710.1| BY-2 kinesin-like protein 10 [Nicotiana tabacum]
Length = 703
Score = 135 bits (339), Expect = 5e-31
Identities = 68/77 (88%), Positives = 73/77 (94%)
Frame = -1
Query: 565 EEDLVNAHRTQVEETMNIVREEMNLLVEADQPGNQLDGYVTRLNAILSQKAAGILQLQTR 386
EEDLVNAHR QVEETM+IV+EEMNLLVEAD PGNQLD Y++RLNAILSQKAAGILQLQ +
Sbjct: 625 EEDLVNAHRKQVEETMDIVKEEMNLLVEADLPGNQLDDYISRLNAILSQKAAGILQLQNQ 684
Query: 385 LAHFQKRLKEHNVLVSS 335
LAHFQKRLKEHNVLVSS
Sbjct: 685 LAHFQKRLKEHNVLVSS 701
>dbj|BAB19066.1| kinesin-like protein [Oryza sativa (japonica cultivar-group)]
Length = 707
Score = 116 bits (291), Expect = 2e-25
Identities = 56/75 (74%), Positives = 68/75 (90%)
Frame = -1
Query: 565 EEDLVNAHRTQVEETMNIVREEMNLLVEADQPGNQLDGYVTRLNAILSQKAAGILQLQTR 386
EEDLV+AHR QVEET+++++EEMNLLVEADQPGNQLD Y+TRL+ ILSQKAAGI+ LQ R
Sbjct: 628 EEDLVSAHRKQVEETLDMIKEEMNLLVEADQPGNQLDDYITRLSGILSQKAAGIVDLQAR 687
Query: 385 LAHFQKRLKEHNVLV 341
LA FQ+RL E+NVL+
Sbjct: 688 LAQFQRRLNENNVLL 702
>gb|AAK70904.1|AC087551_3 kinesin-like protein [Oryza sativa]
Length = 800
Score = 96.3 bits (238), Expect = 2e-19
Identities = 46/74 (62%), Positives = 58/74 (78%)
Frame = -1
Query: 565 EEDLVNAHRTQVEETMNIVREEMNLLVEADQPGNQLDGYVTRLNAILSQKAAGILQLQTR 386
EE L+ AHR ++E TM IVREEMNLL E DQPG+ +D YVT+L+ +LS+KAAG++ LQ R
Sbjct: 720 EEALIAAHRKEIENTMEIVREEMNLLAEVDQPGSLIDNYVTQLSFLLSRKAAGLVSLQAR 779
Query: 385 LAHFQKRLKEHNVL 344
LA FQ RLKE +L
Sbjct: 780 LARFQHRLKEQEIL 793
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 426,369,228
Number of Sequences: 1393205
Number of extensions: 8279426
Number of successful extensions: 19120
Number of sequences better than 10.0: 48
Number of HSP's better than 10.0 without gapping: 18659
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 19120
length of database: 448,689,247
effective HSP length: 116
effective length of database: 287,077,467
effective search space used: 20382500157
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)