Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC005592A_C01 KMC005592A_c01
(672 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
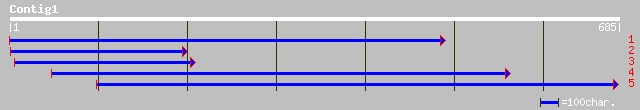
ref|NP_567059.1| putative protein; protein id: At3g57890.1, supp... 132 3e-30
pir||T46011 hypothetical protein T10K17.100 - Arabidopsis thalia... 114 2e-24
ref|NP_181752.1| hypothetical protein; protein id: At2g42230.1 [... 99 5e-20
ref|XP_148441.2| RIKEN cDNA 5730478M09 [Mus musculus] gi|2633351... 39 0.064
ref|NP_060608.1| hypothetical protein FLJ10560 [Homo sapiens] gi... 39 0.084
>ref|NP_567059.1| putative protein; protein id: At3g57890.1, supported by cDNA:
gi_15450720, supported by cDNA: gi_17380619 [Arabidopsis
thaliana] gi|15450721|gb|AAK96632.1|
AT3g57890/T10K17_100 [Arabidopsis thaliana]
gi|17380620|gb|AAL36073.1| AT3g57890/T10K17_100
[Arabidopsis thaliana]
Length = 573
Score = 132 bits (333), Expect = 3e-30
Identities = 63/99 (63%), Positives = 74/99 (74%)
Frame = -2
Query: 671 QAESATQLDPDQFTNFVIPDWIAGDSTVTAKHHPFTLPDVYVASQHRNPTHLGEIRKLLP 492
QAESA +DPDQF NF+IP+W +G + K +PF LPD Y+A+Q RN + E R+ L
Sbjct: 475 QAESAASVDPDQFVNFLIPNWFSGAELGSTKDNPFPLPDAYMAAQQRNLKNFEETRRSLK 534
Query: 491 EASLEESRTREFSSALHVYFKDWLYASGYIRQLYCLPGD 375
EA LEE+R RE SSALHVYFKDWLYASG IRQLYCL GD
Sbjct: 535 EAPLEENRKRELSSALHVYFKDWLYASGNIRQLYCLQGD 573
>pir||T46011 hypothetical protein T10K17.100 - Arabidopsis thaliana
gi|6729532|emb|CAB67617.1| putative protein [Arabidopsis
thaliana]
Length = 570
Score = 114 bits (284), Expect = 2e-24
Identities = 60/112 (53%), Positives = 70/112 (61%), Gaps = 13/112 (11%)
Frame = -2
Query: 671 QAESATQLDPDQ-------------FTNFVIPDWIAGDSTVTAKHHPFTLPDVYVASQHR 531
QAESA +DPD T IP+W +G + K +PF LPD Y+A+Q R
Sbjct: 459 QAESAASVDPDHVMKHLMTCFSVVSMTMAQIPNWFSGAELGSTKDNPFPLPDAYMAAQQR 518
Query: 530 NPTHLGEIRKLLPEASLEESRTREFSSALHVYFKDWLYASGYIRQLYCLPGD 375
N + E R+ L EA LEE+R RE SSALHVYFKDWLYASG IRQLYCL GD
Sbjct: 519 NLKNFEETRRSLKEAPLEENRKRELSSALHVYFKDWLYASGNIRQLYCLQGD 570
>ref|NP_181752.1| hypothetical protein; protein id: At2g42230.1 [Arabidopsis
thaliana] gi|7452467|pir||T00932 hypothetical protein
At2g42230 [imported] - Arabidopsis thaliana
gi|2673914|gb|AAB88648.1| hypothetical protein
[Arabidopsis thaliana]
Length = 551
Score = 99.0 bits (245), Expect = 5e-20
Identities = 46/85 (54%), Positives = 60/85 (70%)
Frame = -2
Query: 671 QAESATQLDPDQFTNFVIPDWIAGDSTVTAKHHPFTLPDVYVASQHRNPTHLGEIRKLLP 492
QAE A+ +DPDQFT F+IP+W AG++ + K +PF LPD Y A Q N +L E R+ L
Sbjct: 418 QAEPASCVDPDQFTTFLIPNWFAGEAIGSTKDNPFPLPDAYKAVQQTNLKNLEETRQSLR 477
Query: 491 EASLEESRTREFSSALHVYFKDWLY 417
E LEE+R RE ++A H+YFKDWLY
Sbjct: 478 ETPLEENRKRELTTAFHMYFKDWLY 502
>ref|XP_148441.2| RIKEN cDNA 5730478M09 [Mus musculus] gi|26333517|dbj|BAC30476.1|
unnamed protein product [Mus musculus]
Length = 475
Score = 38.9 bits (89), Expect = 0.064
Identities = 24/86 (27%), Positives = 43/86 (49%)
Frame = -2
Query: 650 LDPDQFTNFVIPDWIAGDSTVTAKHHPFTLPDVYVASQHRNPTHLGEIRKLLPEASLEES 471
L P +F FVIP + GD+ P LP Y + + + +K + EA L +
Sbjct: 384 LPPSEFYIFVIPFEMEGDTA----EIPGGLPSAYQKALAQREESIHIWQKTVKEARLTKE 439
Query: 470 RTREFSSALHVYFKDWLYASGYIRQL 393
+ ++F + + F +WL ++G+ +QL
Sbjct: 440 QRKQFQALVENKFYEWLVSTGHRQQL 465
>ref|NP_060608.1| hypothetical protein FLJ10560 [Homo sapiens]
gi|7022670|dbj|BAA91682.1| unnamed protein product [Homo
sapiens] gi|19343947|gb|AAH25748.1| hypothetical protein
FLJ10560 [Homo sapiens]
Length = 557
Score = 38.5 bits (88), Expect = 0.084
Identities = 25/86 (29%), Positives = 43/86 (49%)
Frame = -2
Query: 650 LDPDQFTNFVIPDWIAGDSTVTAKHHPFTLPDVYVASQHRNPTHLGEIRKLLPEASLEES 471
L P +F F+IP + GD+T P LP VY + + + +K + EA L +
Sbjct: 461 LPPCEFYVFIIPFEMEGDTT----EIPGGLPSVYQKALGQREQKIQIWQKTVKEAHLTKD 516
Query: 470 RTREFSSALHVYFKDWLYASGYIRQL 393
+ ++F + F +WL +G+ +QL
Sbjct: 517 QRKQFQVLVENKFYEWLINTGHRQQL 542
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 592,414,210
Number of Sequences: 1393205
Number of extensions: 12699150
Number of successful extensions: 33049
Number of sequences better than 10.0: 13
Number of HSP's better than 10.0 without gapping: 31708
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 33040
length of database: 448,689,247
effective HSP length: 119
effective length of database: 282,897,852
effective search space used: 29421376608
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)