Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC005570A_C01 KMC005570A_c01
(817 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
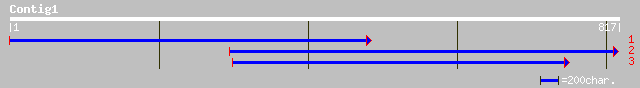
ref|NP_180512.1| unknown protein; protein id: At2g29510.1, suppo... 55 1e-06
ref|NP_200710.1| putative protein; protein id: At5g59020.1 [Arab... 48 2e-04
ref|NP_196868.1| putative protein; protein id: At5g13640.1, supp... 40 0.041
ref|NP_195723.1| putative protein; protein id: At5g01030.1, supp... 39 0.12
ref|NP_181329.1| hypothetical protein; protein id: At2g37930.1 [... 34 0.35
>ref|NP_180512.1| unknown protein; protein id: At2g29510.1, supported by cDNA:
gi_20260411 [Arabidopsis thaliana]
gi|25408032|pir||C84697 hypothetical protein At2g29510
[imported] - Arabidopsis thaliana
gi|3980384|gb|AAC95187.1| unknown protein [Arabidopsis
thaliana] gi|20260412|gb|AAM13104.1| unknown protein
[Arabidopsis thaliana]
Length = 839
Score = 55.1 bits (131), Expect = 1e-06
Identities = 24/55 (43%), Positives = 39/55 (70%)
Frame = -2
Query: 804 ADPFELFLQGNDQDQDNGPTFKFSLFKPGVYSVAFDSSLSHLPAFFICLALVDGK 640
+D F+LF QG Q+ +N P F+ ++ GVY+V +++SLS L AF IC+A+ +G+
Sbjct: 725 SDAFKLFFQGGVQENNNQPYLSFTTYREGVYAVEYNTSLSLLQAFSICIAVNEGR 779
>ref|NP_200710.1| putative protein; protein id: At5g59020.1 [Arabidopsis thaliana]
gi|10177630|dbj|BAB10778.1|
gb|AAC95187.1~gene_id:K18B18.1~similar to unknown
protein [Arabidopsis thaliana]
Length = 780
Score = 47.8 bits (112), Expect = 2e-04
Identities = 24/52 (46%), Positives = 32/52 (61%)
Frame = -2
Query: 795 FELFLQGNDQDQDNGPTFKFSLFKPGVYSVAFDSSLSHLPAFFICLALVDGK 640
FELF G ++ P F K G+YSVA++SSLS L AF IC+AL + +
Sbjct: 682 FELFFLGEQAEEH--PFLSFKPIKEGIYSVAYNSSLSQLQAFSICMALAESR 731
>ref|NP_196868.1| putative protein; protein id: At5g13640.1, supported by cDNA:
gi_15450694 [Arabidopsis thaliana]
gi|9758029|dbj|BAB08690.1| contains similarity to
unknown protein~gene_id:MSH12.10~ref|NP_014405.1
[Arabidopsis thaliana] gi|15450695|gb|AAK96619.1|
AT5g13640/MSH12_10 [Arabidopsis thaliana]
Length = 671
Score = 40.0 bits (92), Expect = 0.041
Identities = 25/78 (32%), Positives = 39/78 (49%), Gaps = 2/78 (2%)
Frame = -2
Query: 798 PFELFLQGNDQDQDNGPTFKFSLFKPGVYSVAFDSSLSHLPAFFICLALVDGKILYAPSG 619
PF++F +++D+D S K GVY+V D ++ L A ++C GK + PSG
Sbjct: 547 PFQIFTSAHEEDED-------SCLKAGVYNVDGDETVPVLSAGYMCAKAWRGKTRFNPSG 599
Query: 618 SRYTIEGKN--PQGNCLD 571
+ I N P N L+
Sbjct: 600 IKTYIREYNHSPPANLLE 617
>ref|NP_195723.1| putative protein; protein id: At5g01030.1, supported by cDNA:
gi_20465614 [Arabidopsis thaliana]
gi|11357833|pir||T45943 hypothetical protein F7J8.10 -
Arabidopsis thaliana gi|6759426|emb|CAB69831.1| putative
protein [Arabidopsis thaliana]
gi|20465615|gb|AAM20139.1| unknown protein [Arabidopsis
thaliana] gi|26983886|gb|AAN86195.1| unknown protein
[Arabidopsis thaliana]
Length = 744
Score = 38.5 bits (88), Expect = 0.12
Identities = 20/53 (37%), Positives = 29/53 (53%)
Frame = -2
Query: 807 FADPFELFLQGNDQDQDNGPTFKFSLFKPGVYSVAFDSSLSHLPAFFICLALV 649
F F LF Q +QD+ P + K G+Y V F S +S L AFF+C+ ++
Sbjct: 667 FNQSFTLFDQ-EVSEQDSSPALAMTELKTGIYRVEFGSFVSPLQAFFVCVTVL 718
>ref|NP_181329.1| hypothetical protein; protein id: At2g37930.1 [Arabidopsis
thaliana] gi|25408578|pir||H84798 hypothetical protein
At2g37930 [imported] - Arabidopsis thaliana
Length = 571
Score = 33.9 bits (76), Expect(2) = 0.35
Identities = 22/48 (45%), Positives = 29/48 (59%)
Frame = -2
Query: 795 FELFLQGNDQDQDNGPTFKFSLFKPGVYSVAFDSSLSHLPAFFICLAL 652
F+LF DQ++D P FK ++SV F SS+S L AFFI LA+
Sbjct: 486 FQLF----DQERDE-PAFKMVSHGDELHSVEFGSSISLLEAFFISLAV 528
Score = 21.9 bits (45), Expect(2) = 0.35
Identities = 8/14 (57%), Positives = 9/14 (64%)
Frame = -3
Query: 524 ASYVAYPPHSPVGR 483
A Y PP SP+GR
Sbjct: 557 AKYATNPPVSPIGR 570
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 653,794,937
Number of Sequences: 1393205
Number of extensions: 13556597
Number of successful extensions: 27124
Number of sequences better than 10.0: 13
Number of HSP's better than 10.0 without gapping: 26279
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 27116
length of database: 448,689,247
effective HSP length: 121
effective length of database: 280,111,442
effective search space used: 42016716300
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)