Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC005567A_C03 KMC005567A_c03
(617 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
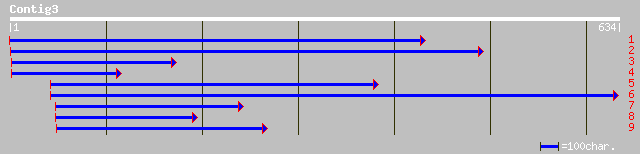
ref|NP_191417.1| putative protein; protein id: At3g58580.1 [Arab... 193 1e-48
pir||T45676 hypothetical protein F14P22.150 - Arabidopsis thalia... 187 7e-47
ref|NP_191415.2| putative protein; protein id: At3g58560.1, supp... 187 7e-47
ref|NP_703540.1| hypothetical protein [Plasmodium falciparum 3D7... 65 7e-10
gb|AAL39488.1| LD05405p [Drosophila melanogaster] 58 1e-07
>ref|NP_191417.1| putative protein; protein id: At3g58580.1 [Arabidopsis thaliana]
gi|11282424|pir||T45678 hypothetical protein F14P22.170
- Arabidopsis thaliana gi|6735375|emb|CAB68196.1|
putative protein [Arabidopsis thaliana]
Length = 605
Score = 193 bits (491), Expect = 1e-48
Identities = 97/125 (77%), Positives = 108/125 (85%), Gaps = 2/125 (1%)
Frame = -2
Query: 616 PSHPDLAIDPLSILRPHSKLVHQSPLVSAYSSFART--IGLGYEQHKRRLDSATNEPLFT 443
P HPDLA+DPL+ILRPH+KL HQ PLVSAYSSF R +GLG EQH+RR+D TNEPLFT
Sbjct: 481 PMHPDLAVDPLNILRPHTKLTHQLPLVSAYSSFVRKGIMGLGLEQHRRRIDLNTNEPLFT 540
Query: 442 NVTRDFIGALDYIFYTADSLVVESLLELLDEESLRKDTALPSPEWSSDHIALLAEFRCCK 263
N TRDFIG DYIFYTAD+L+VESLLELLDE+ LRKDTALPSPEWSS+HIALLAEFRC
Sbjct: 541 NCTRDFIGTHDYIFYTADTLMVESLLELLDEDGLRKDTALPSPEWSSNHIALLAEFRCTP 600
Query: 262 NRSRR 248
R+RR
Sbjct: 601 -RTRR 604
>pir||T45676 hypothetical protein F14P22.150 - Arabidopsis thaliana
gi|6735373|emb|CAB68194.1| putative protein [Arabidopsis
thaliana]
Length = 597
Score = 187 bits (476), Expect = 7e-47
Identities = 97/124 (78%), Positives = 105/124 (84%), Gaps = 1/124 (0%)
Frame = -2
Query: 616 PSHPDLAIDPLSILRPHSKLVHQSPLVSAYSSFARTIG-LGYEQHKRRLDSATNEPLFTN 440
P HPDL +DPL ILRPHSKL HQ PLVSAYS FA+ G + EQ +RRLD A++EPLFTN
Sbjct: 471 PLHPDLMVDPLGILRPHSKLTHQLPLVSAYSQFAKMGGNVITEQQRRRLDPASSEPLFTN 530
Query: 439 VTRDFIGALDYIFYTADSLVVESLLELLDEESLRKDTALPSPEWSSDHIALLAEFRCCKN 260
TRDFIG LDYIFYTAD+L VESLLELLDEESLRKDTALPSPEWSSDHIALLAEFRC
Sbjct: 531 CTRDFIGTLDYIFYTADTLTVESLLELLDEESLRKDTALPSPEWSSDHIALLAEFRCMP- 589
Query: 259 RSRR 248
R+RR
Sbjct: 590 RARR 593
>ref|NP_191415.2| putative protein; protein id: At3g58560.1, supported by cDNA:
gi_18087603 [Arabidopsis thaliana]
gi|18087604|gb|AAL58932.1|AF462845_1
AT3g58560/F14P22_150 [Arabidopsis thaliana]
gi|22137046|gb|AAM91368.1| At3g58560/F14P22_150
[Arabidopsis thaliana]
Length = 602
Score = 187 bits (476), Expect = 7e-47
Identities = 97/124 (78%), Positives = 105/124 (84%), Gaps = 1/124 (0%)
Frame = -2
Query: 616 PSHPDLAIDPLSILRPHSKLVHQSPLVSAYSSFARTIG-LGYEQHKRRLDSATNEPLFTN 440
P HPDL +DPL ILRPHSKL HQ PLVSAYS FA+ G + EQ +RRLD A++EPLFTN
Sbjct: 476 PLHPDLMVDPLGILRPHSKLTHQLPLVSAYSQFAKMGGNVITEQQRRRLDPASSEPLFTN 535
Query: 439 VTRDFIGALDYIFYTADSLVVESLLELLDEESLRKDTALPSPEWSSDHIALLAEFRCCKN 260
TRDFIG LDYIFYTAD+L VESLLELLDEESLRKDTALPSPEWSSDHIALLAEFRC
Sbjct: 536 CTRDFIGTLDYIFYTADTLTVESLLELLDEESLRKDTALPSPEWSSDHIALLAEFRCMP- 594
Query: 259 RSRR 248
R+RR
Sbjct: 595 RARR 598
>ref|NP_703540.1| hypothetical protein [Plasmodium falciparum 3D7]
gi|23504682|emb|CAD51560.1| hypothetical protein
[Plasmodium falciparum 3D7]
Length = 2488
Score = 65.1 bits (157), Expect = 7e-10
Identities = 48/122 (39%), Positives = 64/122 (52%), Gaps = 9/122 (7%)
Frame = -2
Query: 613 SHPDLAIDPLSILRPHSKLVHQSPLVSAYS-SFARTIGLGYEQHKRRLDSATNEPLFTNV 437
+H D D S+L KL H L SAY+ S + L E++ + EPLFTN
Sbjct: 2366 THEDFNSDKYSLLTD-LKLGHNLNLKSAYAISKLLSQKLNPEEYN---NLELYEPLFTNY 2421
Query: 436 TRDFIGALDYIFYTADSLVVESLLELLDEESL--------RKDTALPSPEWSSDHIALLA 281
T +FIG LDYIFY ++L + S + + DE L D ALPSP SDH+ L+A
Sbjct: 2422 TSNFIGCLDYIFYNDENLNIISTVNVADENQLIQEAQMYQLSDCALPSPIRPSDHLPLIA 2481
Query: 280 EF 275
+F
Sbjct: 2482 QF 2483
>gb|AAL39488.1| LD05405p [Drosophila melanogaster]
Length = 526
Score = 57.8 bits (138), Expect = 1e-07
Identities = 28/69 (40%), Positives = 43/69 (61%)
Frame = -2
Query: 478 RLDSATNEPLFTNVTRDFIGALDYIFYTADSLVVESLLELLDEESLRKDTALPSPEWSSD 299
++ SA P +T+ T F G LDY+FY D V ++ L EE L+ +TA+PS + SD
Sbjct: 454 KMASAYGAPEYTHYTTLFAGCLDYVFYQNDRFEVLKVVPLPTEEELKANTAIPSAVFPSD 513
Query: 298 HIALLAEFR 272
H+AL+A+ +
Sbjct: 514 HVALVADLK 522
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 512,582,763
Number of Sequences: 1393205
Number of extensions: 10466800
Number of successful extensions: 34856
Number of sequences better than 10.0: 89
Number of HSP's better than 10.0 without gapping: 33508
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 34749
length of database: 448,689,247
effective HSP length: 118
effective length of database: 284,291,057
effective search space used: 24733321959
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)