Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC005547A_C01 KMC005547A_c01
(633 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
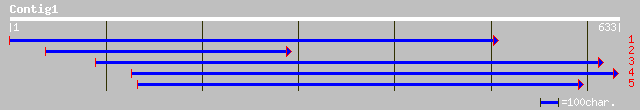
ref|NP_568874.1| non phototropic hypocotyl 1-like; protein id: A... 145 4e-34
gb|AAB39188.1| putative serine/threonine protein kinase [Arabido... 145 4e-34
pir||S42868 serine/threonine protein kinase (EC 2.7.1.-), nonpho... 129 3e-29
dbj|BAA84779.1| nonphototrophic hypocotyl 1b [Oryza sativa (japo... 112 4e-24
dbj|BAA84780.1| nonphototrophic hypocotyl 1a [Oryza sativa (japo... 94 1e-18
>ref|NP_568874.1| non phototropic hypocotyl 1-like; protein id: At5g58140.1, supported
by cDNA: gi_5391441 [Arabidopsis thaliana]
gi|11358837|pir||T51600 serine/threonine protein kinase
(EC 2.7.1.-), nonphototropic hypocotyl protein 1-like
[similarity] - Arabidopsis thaliana
gi|5391442|gb|AAC27293.2| non phototropic hypocotyl
1-like [Arabidopsis thaliana]
Length = 915
Score = 145 bits (366), Expect = 4e-34
Identities = 70/90 (77%), Positives = 75/90 (82%)
Frame = -3
Query: 631 FPSSIPVSLAARQLINALLQRDPASRLGSTTGANEIKQHPFFREINWPLIRNMSPPPLDV 452
FPSSIPVSL RQLIN LL RDP+SRLGS GANEIKQH FFR INWPLIR MSPPPLD
Sbjct: 823 FPSSIPVSLVGRQLINTLLNRDPSSRLGSKGGANEIKQHAFFRGINWPLIRGMSPPPLDA 882
Query: 451 PLQLIGKDPVAKNINWEDDGVLVSSVDMDI 362
PL +I KDP AK+I WEDDGVLV+S D+DI
Sbjct: 883 PLSIIEKDPNAKDIKWEDDGVLVNSTDLDI 912
>gb|AAB39188.1| putative serine/threonine protein kinase [Arabidopsis thaliana]
Length = 356
Score = 145 bits (366), Expect = 4e-34
Identities = 70/90 (77%), Positives = 75/90 (82%)
Frame = -3
Query: 631 FPSSIPVSLAARQLINALLQRDPASRLGSTTGANEIKQHPFFREINWPLIRNMSPPPLDV 452
FPSSIPVSL RQLIN LL RDP+SRLGS GANEIKQH FFR INWPLIR MSPPPLD
Sbjct: 264 FPSSIPVSLVGRQLINTLLNRDPSSRLGSKGGANEIKQHAFFRGINWPLIRGMSPPPLDA 323
Query: 451 PLQLIGKDPVAKNINWEDDGVLVSSVDMDI 362
PL +I KDP AK+I WEDDGVLV+S D+DI
Sbjct: 324 PLSIIEKDPNAKDIKWEDDGVLVNSTDLDI 353
>pir||S42868 serine/threonine protein kinase (EC 2.7.1.-), nonphototropic
hypocotyl protein 1-like [similarity] - spinach
(fragment) gi|457711|emb|CAA82993.1| protein kinase
[Spinacia oleracea]
Length = 724
Score = 129 bits (324), Expect = 3e-29
Identities = 60/87 (68%), Positives = 72/87 (81%)
Frame = -3
Query: 631 FPSSIPVSLAARQLINALLQRDPASRLGSTTGANEIKQHPFFREINWPLIRNMSPPPLDV 452
FPSSIPVSL+ARQLI ALL RDPA+RLG+ GA+EIK+HP+FR INWPLIR M PP L+
Sbjct: 636 FPSSIPVSLSARQLIYALLNRDPATRLGTQGGASEIKEHPYFRGINWPLIRCMDPPTLEA 695
Query: 451 PLQLIGKDPVAKNINWEDDGVLVSSVD 371
P +LIG+DP AK + W+DDGVL S +D
Sbjct: 696 PFKLIGRDPNAKEVKWDDDGVLASPMD 722
>dbj|BAA84779.1| nonphototrophic hypocotyl 1b [Oryza sativa (japonica cultivar-group)]
Length = 907
Score = 112 bits (280), Expect = 4e-24
Identities = 53/72 (73%), Positives = 63/72 (86%)
Frame = -3
Query: 631 FPSSIPVSLAARQLINALLQRDPASRLGSTTGANEIKQHPFFREINWPLIRNMSPPPLDV 452
FPSSIPVSLAA+QLI+ LLQRDP++R+GS GAN+IKQH FF++INWPLIR MSPP LDV
Sbjct: 822 FPSSIPVSLAAKQLIHGLLQRDPSNRIGSNAGANDIKQHSFFQDINWPLIRCMSPPELDV 881
Query: 451 PLQLIGKDPVAK 416
PL+LIGK+ K
Sbjct: 882 PLKLIGKETQPK 893
>dbj|BAA84780.1| nonphototrophic hypocotyl 1a [Oryza sativa (japonica cultivar-group)]
Length = 921
Score = 94.0 bits (232), Expect = 1e-18
Identities = 48/72 (66%), Positives = 55/72 (75%)
Frame = -3
Query: 631 FPSSIPVSLAARQLINALLQRDPASRLGSTTGANEIKQHPFFREINWPLIRNMSPPPLDV 452
FP+SI VSLAARQL+ LL RDPA+RLGS GANEIK HPFFR INWPLIR +PP L++
Sbjct: 840 FPASISVSLAARQLMYRLLHRDPANRLGSYEGANEIKGHPFFRGINWPLIRATAPPKLEI 899
Query: 451 PLQLIGKDPVAK 416
P L KD + K
Sbjct: 900 P--LFSKDDMEK 909
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 531,529,703
Number of Sequences: 1393205
Number of extensions: 11006700
Number of successful extensions: 25622
Number of sequences better than 10.0: 794
Number of HSP's better than 10.0 without gapping: 24711
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 25326
length of database: 448,689,247
effective HSP length: 118
effective length of database: 284,291,057
effective search space used: 26154777244
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)