Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KMC005512A_C01 KMC005512A_c01
(635 letters)
Database: nr
1,393,205 sequences; 448,689,247 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
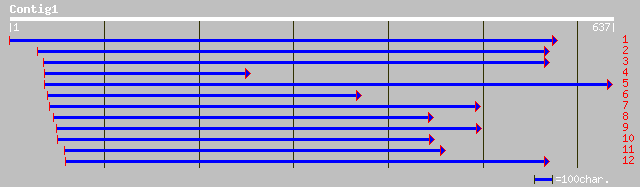
gb|AAO27794.1| glycosyl hydrolase [Gossypium hirsutum] 146 2e-34
gb|AAK84953.2| putative beta-mannosidase [Gossypium hirsutum] 146 2e-34
ref|NP_563833.1| expressed protein; protein id: At1g09010.1, sup... 111 9e-24
pir||A86222 hypothetical protein [imported] - Arabidopsis thalia... 84 2e-15
ref|NP_643619.1| glycosyl hydrolase [Xanthomonas axonopodis pv. ... 44 0.002
>gb|AAO27794.1| glycosyl hydrolase [Gossypium hirsutum]
Length = 976
Score = 146 bits (369), Expect = 2e-34
Identities = 75/155 (48%), Positives = 105/155 (67%), Gaps = 8/155 (5%)
Frame = -2
Query: 631 LEESTYHIQVHVTNTSKRPDSESSTFEHSSRAMLDSMETVHSGAAKAHDR--------GW 476
++ S+Y +++ V N SK+PD ++ T++++ D + + D G
Sbjct: 818 IKGSSYEVEMKVLNKSKKPDPKTLTYKNNFAVRNDDSDFDMTSLKPIPDTRTDLKQPTGL 877
Query: 475 FKRIIHRCFAGKSDGLKVCEIDGHDVGVAFFLHFSVHTSKKDYKEGEDTRILPVHYSDNY 296
F+R+ +R F+ +SDGL+V EI+G D GVAFFL+FSVH +K +++EGED+RILPVHYSDNY
Sbjct: 878 FQRL-YRQFSRESDGLRVAEINGSDGGVAFFLNFSVHGAKLEHEEGEDSRILPVHYSDNY 936
Query: 295 FSLVPGETMPVNISFDVPQGVTPRVVLHGWNYEGG 191
FSLVPGE M + ISF VP GV+PRV L GWNY G
Sbjct: 937 FSLVPGEEMSIKISFKVPPGVSPRVTLRGWNYHHG 971
>gb|AAK84953.2| putative beta-mannosidase [Gossypium hirsutum]
Length = 786
Score = 146 bits (369), Expect = 2e-34
Identities = 75/155 (48%), Positives = 105/155 (67%), Gaps = 8/155 (5%)
Frame = -2
Query: 631 LEESTYHIQVHVTNTSKRPDSESSTFEHSSRAMLDSMETVHSGAAKAHDR--------GW 476
++ S+Y +++ V N SK+PD ++ T++++ D + + D G
Sbjct: 628 IKGSSYEVEMKVLNKSKKPDPKTLTYKNNFAVRNDDSDFDMTSLKPIPDTRTDLKQPTGL 687
Query: 475 FKRIIHRCFAGKSDGLKVCEIDGHDVGVAFFLHFSVHTSKKDYKEGEDTRILPVHYSDNY 296
F+R+ +R F+ +SDGL+V EI+G D GVAFFL+FSVH +K +++EGED+RILPVHYSDNY
Sbjct: 688 FQRL-YRQFSRESDGLRVAEINGSDGGVAFFLNFSVHGAKLEHEEGEDSRILPVHYSDNY 746
Query: 295 FSLVPGETMPVNISFDVPQGVTPRVVLHGWNYEGG 191
FSLVPGE M + ISF VP GV+PRV L GWNY G
Sbjct: 747 FSLVPGEEMSIKISFKVPPGVSPRVTLRGWNYHHG 781
>ref|NP_563833.1| expressed protein; protein id: At1g09010.1, supported by cDNA:
gi_15028222, supported by cDNA: gi_20259144 [Arabidopsis
thaliana]
Length = 647
Score = 111 bits (277), Expect = 9e-24
Identities = 66/145 (45%), Positives = 85/145 (58%), Gaps = 4/145 (2%)
Frame = -2
Query: 622 STYHIQVHVTNTSKRPDSESSTFEHSSRAMLDSMETVHSGAAKAHDRGWFKRIIHRCF-- 449
S Y ++V+V NTS+ +++ E R D G +++ RC
Sbjct: 515 SRYELEVNVHNTSRANLAKNVVQEDEKR-----------------DLGLLQKLFSRCVVS 557
Query: 448 AGKSDGLKVCEIDGHDVGVAFFLHFSVHTSKKDYKEGEDTRILPVHYSDNYFSLVPGETM 269
A + GLKV E+ G D GVAFFL FSVH ++ E +DTRILPVHYSDNYFSLVPGE+M
Sbjct: 558 ADSNRGLKVVEMKGSDSGVAFFLRFSVHNAET---EKQDTRILPVHYSDNYFSLVPGESM 614
Query: 268 PVNISFDVPQGV--TPRVVLHGWNY 200
ISF P G+ +PRV+L GWNY
Sbjct: 615 SFKISFAAPTGMKKSPRVMLQGWNY 639
>pir||A86222 hypothetical protein [imported] - Arabidopsis thaliana
gi|2342683|gb|AAB70407.1| Contains similarity to Bos
beta-mannosidase (gb|U46067). [Arabidopsis thaliana]
Length = 810
Score = 84.0 bits (206), Expect = 2e-15
Identities = 46/91 (50%), Positives = 55/91 (59%)
Frame = -2
Query: 472 KRIIHRCFAGKSDGLKVCEIDGHDVGVAFFLHFSVHTSKKDYKEGEDTRILPVHYSDNYF 293
K ++ + GLKV E+ G D GVAFFL FSVH ++ E +DTRILPVHYSDNYF
Sbjct: 727 KNVVQEDEKHSNRGLKVVEMKGSDSGVAFFLRFSVHNAET---EKQDTRILPVHYSDNYF 783
Query: 292 SLVPGETMPVNISFDVPQGVTPRVVLHGWNY 200
SLVPG +PRV+L GWNY
Sbjct: 784 SLVPG------------MKKSPRVMLQGWNY 802
>ref|NP_643619.1| glycosyl hydrolase [Xanthomonas axonopodis pv. citri str. 306]
gi|21109656|gb|AAM38155.1| glycosyl hydrolase
[Xanthomonas axonopodis pv. citri str. 306]
Length = 891
Score = 43.5 bits (101), Expect = 0.002
Identities = 25/52 (48%), Positives = 30/52 (57%), Gaps = 3/52 (5%)
Frame = -2
Query: 343 EGEDTRILPVHYSDNYFSLVPGETMPVNISFDVPQGVTPR---VVLHGWNYE 197
+ + RILP +YSDNY SLVPGE V+I P T R + L GWN E
Sbjct: 832 DAQGQRILPAYYSDNYLSLVPGEERVVDIR--GPSAATLRNATLQLRGWNAE 881
Database: nr
Posted date: Apr 1, 2003 2:05 AM
Number of letters in database: 448,689,247
Number of sequences in database: 1,393,205
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 572,551,724
Number of Sequences: 1393205
Number of extensions: 12696508
Number of successful extensions: 31453
Number of sequences better than 10.0: 18
Number of HSP's better than 10.0 without gapping: 29893
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 31271
length of database: 448,689,247
effective HSP length: 118
effective length of database: 284,291,057
effective search space used: 26439068301
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)